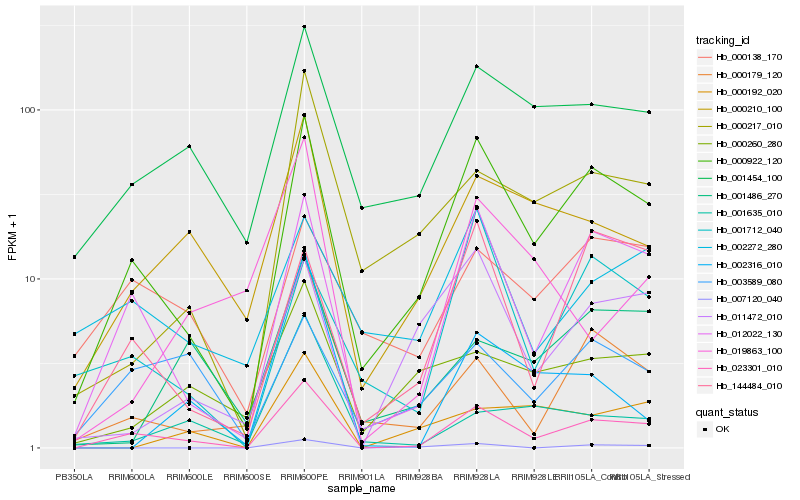
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023301_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 2 |
Hb_000922_120 |
0.1390882928 |
- |
- |
PREDICTED: carbonic anhydrase 2-like [Jatropha curcas] |
| 3 |
Hb_012022_130 |
0.141912053 |
- |
- |
Vacuolar cation/proton exchanger 1a, putative [Ricinus communis] |
| 4 |
Hb_000210_100 |
0.2385818215 |
- |
- |
pepsin A, putative [Ricinus communis] |
| 5 |
Hb_001454_100 |
0.2421854776 |
- |
- |
PREDICTED: protein RALF-like 33 [Jatropha curcas] |
| 6 |
Hb_000179_120 |
0.2434071134 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 7 |
Hb_000217_010 |
0.2548034098 |
- |
- |
hypothetical protein POPTR_0341s00210g [Populus trichocarpa] |
| 8 |
Hb_144484_010 |
0.261818793 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 9 |
Hb_003589_080 |
0.2673181045 |
- |
- |
PREDICTED: uncharacterized protein LOC105107863 [Populus euphratica] |
| 10 |
Hb_000260_280 |
0.2763074482 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 11 |
Hb_000138_170 |
0.2770155578 |
- |
- |
putative plastid-lipid-associated protein, partial [Hevea brasiliensis] |
| 12 |
Hb_007120_040 |
0.2785179726 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000192_020 |
0.2795260703 |
- |
- |
hypothetical protein JCGZ_17351 [Jatropha curcas] |
| 14 |
Hb_011472_010 |
0.2813437972 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 15 |
Hb_001486_270 |
0.2814365088 |
- |
- |
Ann10 [Manihot esculenta] |
| 16 |
Hb_001712_040 |
0.2882787773 |
- |
- |
Calmodulin, putative [Ricinus communis] |
| 17 |
Hb_002272_280 |
0.2897621336 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |
| 18 |
Hb_002316_010 |
0.2898725404 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 19 |
Hb_001635_010 |
0.2967644756 |
- |
- |
- |
| 20 |
Hb_019863_100 |
0.2977469565 |
- |
- |
PREDICTED: alanine--glyoxylate aminotransferase 2 homolog 2, mitochondrial-like [Jatropha curcas] |