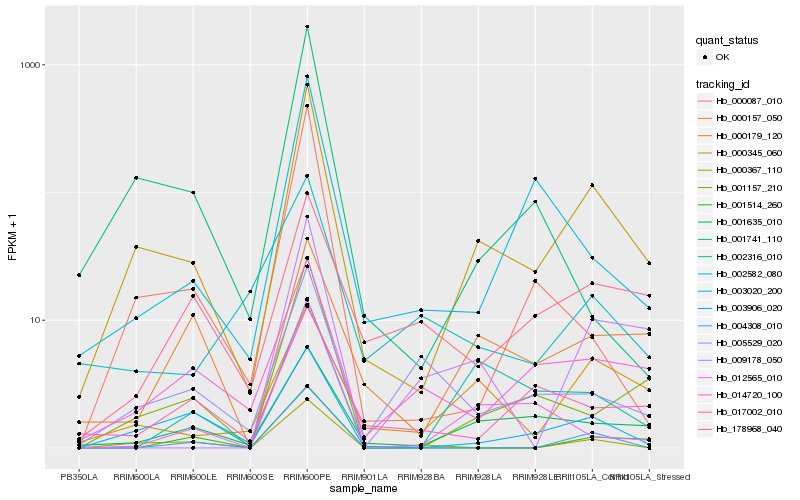
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_060 |
0.0 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 2 |
Hb_003906_020 |
0.2006954202 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 3 |
Hb_001635_010 |
0.2035469797 |
- |
- |
- |
| 4 |
Hb_001157_210 |
0.2139708779 |
- |
- |
hypothetical protein CISIN_1g038260mg [Citrus sinensis] |
| 5 |
Hb_000179_120 |
0.2320371844 |
- |
- |
PREDICTED: corepressor interacting with RBPJ 1-like [Jatropha curcas] |
| 6 |
Hb_014720_100 |
0.238102806 |
- |
- |
PREDICTED: probable boron transporter 2 [Jatropha curcas] |
| 7 |
Hb_000367_110 |
0.2623002163 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 8 |
Hb_003020_200 |
0.2625937082 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 9 |
Hb_012565_010 |
0.2629228282 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 10 |
Hb_002316_010 |
0.2634295239 |
- |
- |
hypothetical protein JCGZ_11498 [Jatropha curcas] |
| 11 |
Hb_009178_050 |
0.2652163485 |
- |
- |
Elongation factor 1-alpha, putative [Ricinus communis] |
| 12 |
Hb_000157_050 |
0.2672168669 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_005529_020 |
0.2684545065 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 14 |
Hb_000087_010 |
0.2702907188 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 15 |
Hb_001741_110 |
0.2706536781 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 16 |
Hb_004308_010 |
0.2727316004 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 17 |
Hb_178968_040 |
0.2731201204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001514_260 |
0.2732257193 |
- |
- |
- |
| 19 |
Hb_017002_010 |
0.27428607 |
- |
- |
PREDICTED: expansin-A15-like [Jatropha curcas] |
| 20 |
Hb_002582_080 |
0.2745742204 |
- |
- |
- |