| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
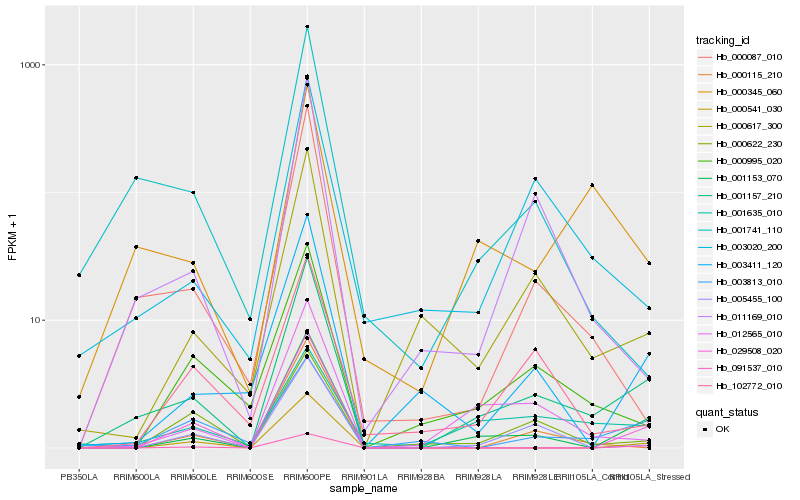
Hb_001157_210 |
0.0 |
- |
- |
hypothetical protein CISIN_1g038260mg [Citrus sinensis] |
| 2 |
Hb_001153_070 |
0.1680667829 |
- |
- |
- |
| 3 |
Hb_012565_010 |
0.2006753424 |
- |
- |
PREDICTED: putative glutamine amidotransferase PB2B2.05 [Jatropha curcas] |
| 4 |
Hb_000617_300 |
0.2064147597 |
- |
- |
RALFL33, putative [Ricinus communis] |
| 5 |
Hb_000087_010 |
0.2069702067 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 6 |
Hb_011169_010 |
0.2136214435 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 7 |
Hb_000345_060 |
0.2139708779 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 8 |
Hb_003411_120 |
0.2163805247 |
- |
- |
hypothetical protein POPTR_0010s13670g [Populus trichocarpa] |
| 9 |
Hb_005455_100 |
0.2215345369 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 10 |
Hb_003020_200 |
0.2218678798 |
- |
- |
PREDICTED: uncharacterized protein LOC105131117 [Populus euphratica] |
| 11 |
Hb_001635_010 |
0.2242680829 |
- |
- |
- |
| 12 |
Hb_001741_110 |
0.2250812259 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 13 |
Hb_000995_020 |
0.2267174187 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_003813_010 |
0.2284894225 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 1-like [Jatropha curcas] |
| 15 |
Hb_102772_010 |
0.2353859037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000622_230 |
0.2362269296 |
- |
- |
PREDICTED: uncharacterized protein LOC105642895 [Jatropha curcas] |
| 17 |
Hb_029508_020 |
0.2365061563 |
- |
- |
- |
| 18 |
Hb_000541_030 |
0.2368896037 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000115_210 |
0.2372965369 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor BEE 3-like [Jatropha curcas] |
| 20 |
Hb_091537_010 |
0.2384314984 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |