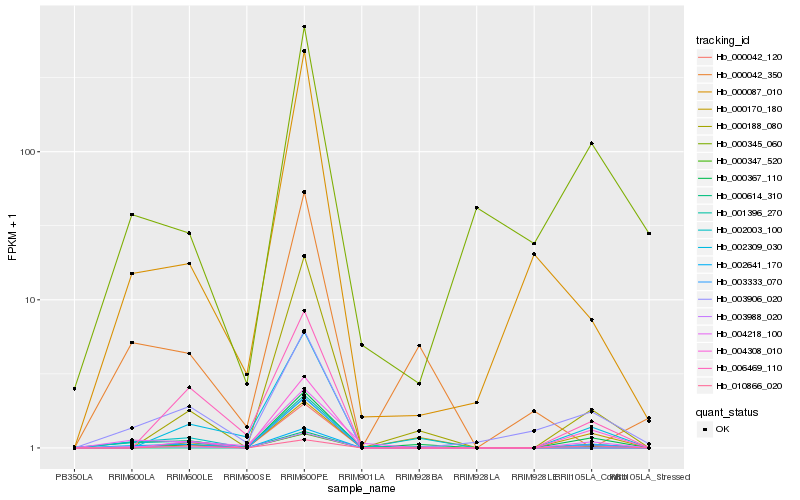
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_110 |
0.0 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 2 |
Hb_002641_170 |
0.1528467361 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_000347_520 |
0.2010688771 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 4 |
Hb_000188_080 |
0.2049729621 |
- |
- |
PREDICTED: uncharacterized protein LOC105629484 [Jatropha curcas] |
| 5 |
Hb_004308_010 |
0.2123268578 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 6 |
Hb_004218_100 |
0.2231615798 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5C [Jatropha curcas] |
| 7 |
Hb_003988_020 |
0.2240471198 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH85-like [Populus euphratica] |
| 8 |
Hb_000170_180 |
0.2279514953 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 9 |
Hb_003333_070 |
0.233822772 |
- |
- |
Ent-kaurenoic acid oxidase, putative [Ricinus communis] |
| 10 |
Hb_003906_020 |
0.2375364237 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 11 |
Hb_002309_030 |
0.2436654436 |
- |
- |
Glycerol dehydrogenase large subunit, putative [Theobroma cacao] |
| 12 |
Hb_010866_020 |
0.2514006717 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_000042_350 |
0.2535215853 |
- |
- |
hypothetical protein JCGZ_00710 [Jatropha curcas] |
| 14 |
Hb_002003_100 |
0.2546831956 |
- |
- |
- |
| 15 |
Hb_001396_270 |
0.2555923189 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 16 |
Hb_000614_310 |
0.2572111742 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000345_060 |
0.2623002163 |
- |
- |
PREDICTED: REF/SRPP-like protein At3g05500 [Jatropha curcas] |
| 18 |
Hb_000087_010 |
0.2647607479 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 19 |
Hb_006469_110 |
0.267732298 |
- |
- |
hypothetical protein JCGZ_12008 [Jatropha curcas] |
| 20 |
Hb_000042_120 |
0.2710706423 |
- |
- |
PREDICTED: uncharacterized protein LOC105632800 [Jatropha curcas] |