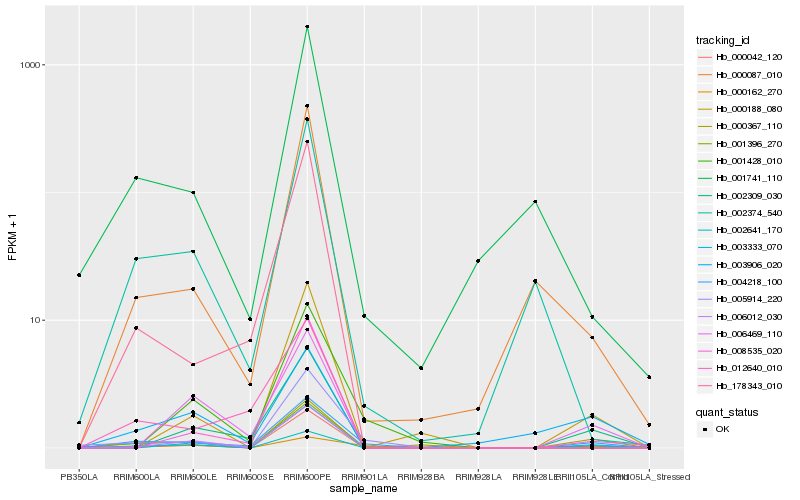
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_100 |
0.0 |
- |
- |
PREDICTED: DNA-directed RNA polymerase V subunit 5C [Jatropha curcas] |
| 2 |
Hb_002641_170 |
0.2057702665 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 3 |
Hb_001396_270 |
0.2078156293 |
- |
- |
4-coumarate--CoA ligase family protein [Populus trichocarpa] |
| 4 |
Hb_000367_110 |
0.2231615798 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 5 |
Hb_005914_220 |
0.2316926873 |
transcription factor |
TF Family: Orphans |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002309_030 |
0.2372332079 |
- |
- |
Glycerol dehydrogenase large subunit, putative [Theobroma cacao] |
| 7 |
Hb_006012_030 |
0.2430500991 |
- |
- |
hypothetical protein POPTR_0008s02380g [Populus trichocarpa] |
| 8 |
Hb_000087_010 |
0.2462891671 |
- |
- |
mta/sah nucleosidase, putative [Ricinus communis] |
| 9 |
Hb_012640_010 |
0.2520578173 |
- |
- |
- |
| 10 |
Hb_001741_110 |
0.252111664 |
rubber biosynthesis |
Gene Name: REF7 |
REF/SRPP-like protein [Glycine soja] |
| 11 |
Hb_002374_540 |
0.2525197238 |
- |
- |
RecName: Full=Esterase; AltName: Full=Early nodule-specific protein homolog; AltName: Full=Latex allergen Hev b 13; AltName: Allergen=Hev b 13; Flags: Precursor [Hevea brasiliensis] |
| 12 |
Hb_178343_010 |
0.2545413092 |
- |
- |
PREDICTED: two-component response regulator ARR17-like [Jatropha curcas] |
| 13 |
Hb_000042_120 |
0.2626360497 |
- |
- |
PREDICTED: uncharacterized protein LOC105632800 [Jatropha curcas] |
| 14 |
Hb_003906_020 |
0.2631572828 |
- |
- |
PREDICTED: uncharacterized protein LOC105648535 [Jatropha curcas] |
| 15 |
Hb_001428_010 |
0.2640183366 |
- |
- |
PREDICTED: transcription factor PRE1-like [Jatropha curcas] |
| 16 |
Hb_003333_070 |
0.2647357223 |
- |
- |
Ent-kaurenoic acid oxidase, putative [Ricinus communis] |
| 17 |
Hb_000162_270 |
0.2675161953 |
- |
- |
PREDICTED: uncharacterized protein LOC104442671 [Eucalyptus grandis] |
| 18 |
Hb_008535_020 |
0.2711638206 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 19 |
Hb_006469_110 |
0.2720572699 |
- |
- |
hypothetical protein JCGZ_12008 [Jatropha curcas] |
| 20 |
Hb_000188_080 |
0.2725683668 |
- |
- |
PREDICTED: uncharacterized protein LOC105629484 [Jatropha curcas] |