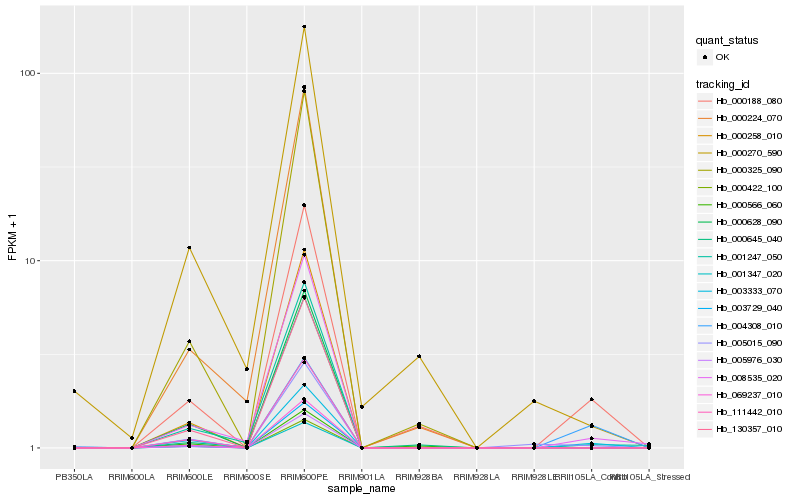
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000188_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629484 [Jatropha curcas] |
| 2 |
Hb_003333_070 |
0.1266439871 |
- |
- |
Ent-kaurenoic acid oxidase, putative [Ricinus communis] |
| 3 |
Hb_008535_020 |
0.1407636572 |
transcription factor |
TF Family: bHLH |
basic helix-loop-helix family protein [Populus trichocarpa] |
| 4 |
Hb_000325_090 |
0.1509208787 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor PRE5-like [Gossypium raimondii] |
| 5 |
Hb_001247_050 |
0.1572117809 |
- |
- |
PREDICTED: WAT1-related protein At1g09380 [Jatropha curcas] |
| 6 |
Hb_004308_010 |
0.1658805809 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 7 |
Hb_000566_060 |
0.1660223451 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003729_040 |
0.1665084398 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 9 |
Hb_069237_010 |
0.1665118108 |
- |
- |
- |
| 10 |
Hb_005015_090 |
0.1665875652 |
- |
- |
PREDICTED: cytochrome P450 78A7 [Jatropha curcas] |
| 11 |
Hb_130357_010 |
0.1666303216 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 12 |
Hb_000270_590 |
0.1671648445 |
- |
- |
hypothetical protein POPTR_0012s07820g [Populus trichocarpa] |
| 13 |
Hb_000258_010 |
0.1671723518 |
- |
- |
lipid transfer precursor protein [Hevea brasiliensis] |
| 14 |
Hb_001347_020 |
0.167391781 |
- |
- |
PREDICTED: cytochrome P450 86B1-like [Populus euphratica] |
| 15 |
Hb_000422_100 |
0.1675265667 |
- |
- |
PREDICTED: TMV resistance protein N-like [Populus euphratica] |
| 16 |
Hb_000645_040 |
0.1675571021 |
transcription factor |
TF Family: GNAT |
N-acetyltransferase, putative [Ricinus communis] |
| 17 |
Hb_005976_030 |
0.1676971775 |
- |
- |
PREDICTED: uncharacterized protein LOC105639715 [Jatropha curcas] |
| 18 |
Hb_111442_010 |
0.1678607194 |
desease resistance |
Gene Name: AAA_16 |
LRR and NB-ARC domains-containing disease resistance protein, putative [Theobroma cacao] |
| 19 |
Hb_000224_070 |
0.1680897121 |
- |
- |
- |
| 20 |
Hb_000628_090 |
0.1686364402 |
- |
- |
hypothetical protein B456_010G019400 [Gossypium raimondii] |