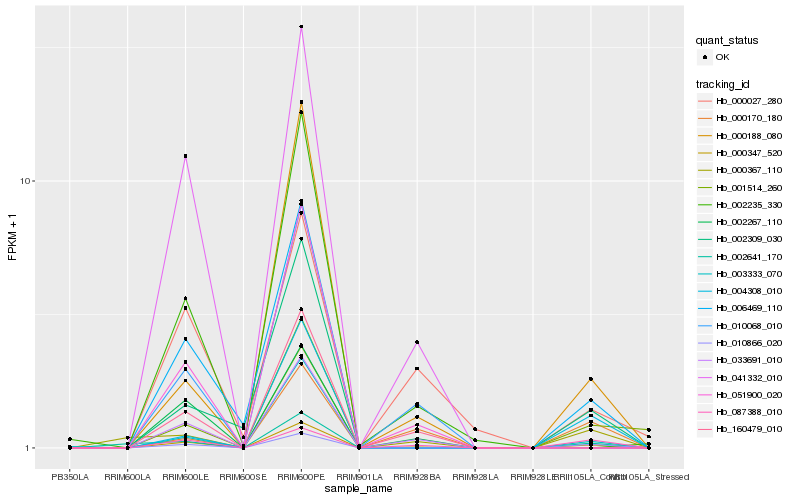
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000347_520 |
0.0 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 2 |
Hb_010866_020 |
0.1499643321 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_006469_110 |
0.1597466728 |
- |
- |
hypothetical protein JCGZ_12008 [Jatropha curcas] |
| 4 |
Hb_003333_070 |
0.1880159976 |
- |
- |
Ent-kaurenoic acid oxidase, putative [Ricinus communis] |
| 5 |
Hb_000188_080 |
0.1969003706 |
- |
- |
PREDICTED: uncharacterized protein LOC105629484 [Jatropha curcas] |
| 6 |
Hb_000367_110 |
0.2010688771 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 7 |
Hb_002309_030 |
0.201580117 |
- |
- |
Glycerol dehydrogenase large subunit, putative [Theobroma cacao] |
| 8 |
Hb_087388_010 |
0.2036518388 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 9 |
Hb_004308_010 |
0.2046356821 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 10 |
Hb_051900_020 |
0.2060342776 |
- |
- |
JHL22C18.7 [Jatropha curcas] |
| 11 |
Hb_033691_010 |
0.2070063625 |
- |
- |
Lignin-forming anionic peroxidase precursor, putative [Ricinus communis] |
| 12 |
Hb_041332_010 |
0.2110740147 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_160479_010 |
0.2125522155 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 14 |
Hb_000027_280 |
0.214939961 |
- |
- |
PREDICTED: uncharacterized protein LOC105642051 [Jatropha curcas] |
| 15 |
Hb_010068_010 |
0.2178753919 |
- |
- |
PREDICTED: uncharacterized protein LOC105633273 [Jatropha curcas] |
| 16 |
Hb_002235_330 |
0.2181320461 |
- |
- |
PREDICTED: expansin-A1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002267_110 |
0.223050295 |
- |
- |
- |
| 18 |
Hb_002641_170 |
0.2268887004 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 19 |
Hb_000170_180 |
0.2273774524 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 20 |
Hb_001514_260 |
0.2286112364 |
- |
- |
- |