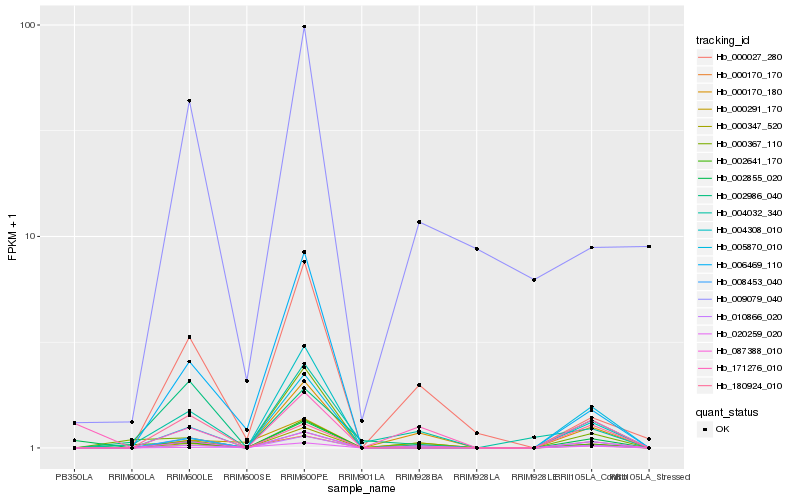
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_087388_010 |
0.0 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 2 |
Hb_008453_040 |
0.0863130391 |
- |
- |
PREDICTED: expansin-A11 [Jatropha curcas] |
| 3 |
Hb_000347_520 |
0.2036518388 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 4 |
Hb_010866_020 |
0.2074481997 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_005870_010 |
0.2107125319 |
- |
- |
- |
| 6 |
Hb_000170_180 |
0.2121204377 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 7 |
Hb_004032_340 |
0.2406012941 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 8 |
Hb_000027_280 |
0.2585895646 |
- |
- |
PREDICTED: uncharacterized protein LOC105642051 [Jatropha curcas] |
| 9 |
Hb_000170_170 |
0.2719715254 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 10 |
Hb_002855_020 |
0.2775874981 |
- |
- |
- |
| 11 |
Hb_002986_040 |
0.2936234276 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 12 |
Hb_000367_110 |
0.2946406903 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |
| 13 |
Hb_000291_170 |
0.2973893025 |
- |
- |
PREDICTED: inactive rhomboid protein 1 isoform X1 [Jatropha curcas] |
| 14 |
Hb_180924_010 |
0.3012673952 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 15 |
Hb_006469_110 |
0.305746775 |
- |
- |
hypothetical protein JCGZ_12008 [Jatropha curcas] |
| 16 |
Hb_171276_010 |
0.3110219382 |
- |
- |
- |
| 17 |
Hb_004308_010 |
0.3154275674 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 18 |
Hb_009079_040 |
0.3229206717 |
- |
- |
BnaA02g30070D [Brassica napus] |
| 19 |
Hb_002641_170 |
0.3246754156 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 20 |
Hb_020259_020 |
0.3247567781 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 4 [Jatropha curcas] |