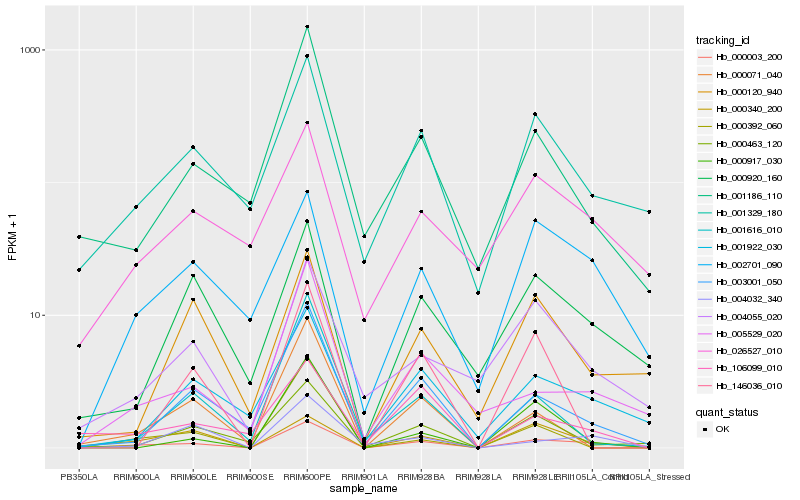
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000003_200 |
0.0 |
- |
- |
hypothetical protein TRIUR3_35397 [Triticum urartu] |
| 2 |
Hb_003001_050 |
0.2209426953 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 3 |
Hb_002701_090 |
0.2328522692 |
- |
- |
hypothetical protein POPTR_0017s13430g [Populus trichocarpa] |
| 4 |
Hb_000340_200 |
0.2350785971 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 5 |
Hb_001922_030 |
0.2360178476 |
- |
- |
PREDICTED: probable membrane-associated kinase regulator 5 [Jatropha curcas] |
| 6 |
Hb_001329_180 |
0.2401688726 |
- |
- |
S-adenosylmethionine synthetase 1 family protein [Populus trichocarpa] |
| 7 |
Hb_004032_340 |
0.2416824309 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 8 |
Hb_005529_020 |
0.249015857 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 9 |
Hb_000463_120 |
0.2500555839 |
- |
- |
LRR receptor-like serine/threonine-protein kinase, putative [Theobroma cacao] |
| 10 |
Hb_004055_020 |
0.2538373383 |
- |
- |
PREDICTED: uncharacterized protein LOC105631433 [Jatropha curcas] |
| 11 |
Hb_000920_160 |
0.2574299953 |
- |
- |
annexin [Manihot esculenta] |
| 12 |
Hb_001186_110 |
0.2597938398 |
- |
- |
hypothetical protein JCGZ_18389 [Jatropha curcas] |
| 13 |
Hb_000917_030 |
0.2600981743 |
- |
- |
PREDICTED: uncharacterized protein LOC102629571 [Citrus sinensis] |
| 14 |
Hb_106099_010 |
0.2625971771 |
- |
- |
- |
| 15 |
Hb_000071_040 |
0.2631207762 |
- |
- |
hypothetical protein POPTR_0001s24580g [Populus trichocarpa] |
| 16 |
Hb_001616_010 |
0.2655340767 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 17 |
Hb_026527_010 |
0.2690217297 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Populus euphratica] |
| 18 |
Hb_146036_010 |
0.2744646565 |
- |
- |
hypothetical protein B456_005G052500 [Gossypium raimondii] |
| 19 |
Hb_000120_940 |
0.2774195076 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000392_060 |
0.2791841128 |
- |
- |
hypothetical protein JCGZ_11629 [Jatropha curcas] |