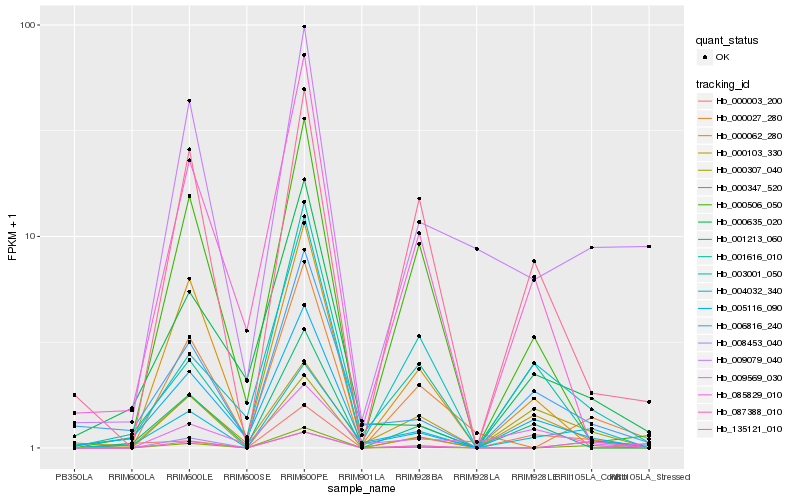
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004032_340 |
0.0 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 2 |
Hb_006816_240 |
0.2030209428 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 3 |
Hb_005116_090 |
0.2047939606 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003001_050 |
0.2283934519 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 5 |
Hb_000103_330 |
0.2327527125 |
- |
- |
PREDICTED: meiotic recombination protein DMC1 homolog isoform X1 [Glycine max] |
| 6 |
Hb_087388_010 |
0.2406012941 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 7 |
Hb_000003_200 |
0.2416824309 |
- |
- |
hypothetical protein TRIUR3_35397 [Triticum urartu] |
| 8 |
Hb_000347_520 |
0.2452680725 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 9 |
Hb_001213_060 |
0.2501070032 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 10 |
Hb_008453_040 |
0.2512510916 |
- |
- |
PREDICTED: expansin-A11 [Jatropha curcas] |
| 11 |
Hb_000635_020 |
0.2535331953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_009569_030 |
0.2536111349 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 21 [Jatropha curcas] |
| 13 |
Hb_001616_010 |
0.257104137 |
- |
- |
hypothetical protein RCOM_1749890 [Ricinus communis] |
| 14 |
Hb_000062_280 |
0.2590761107 |
- |
- |
plant mitotic spindle assembly checkpoint protein mad2, putative [Ricinus communis] |
| 15 |
Hb_000307_040 |
0.2604940155 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 16 |
Hb_009079_040 |
0.2609359732 |
- |
- |
BnaA02g30070D [Brassica napus] |
| 17 |
Hb_135121_010 |
0.2611407833 |
- |
- |
PREDICTED: uncharacterized protein LOC105635544 [Jatropha curcas] |
| 18 |
Hb_000027_280 |
0.2614324105 |
- |
- |
PREDICTED: uncharacterized protein LOC105642051 [Jatropha curcas] |
| 19 |
Hb_085829_010 |
0.261967077 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 20 |
Hb_000506_050 |
0.2623814559 |
- |
- |
PREDICTED: thymidine kinase [Jatropha curcas] |