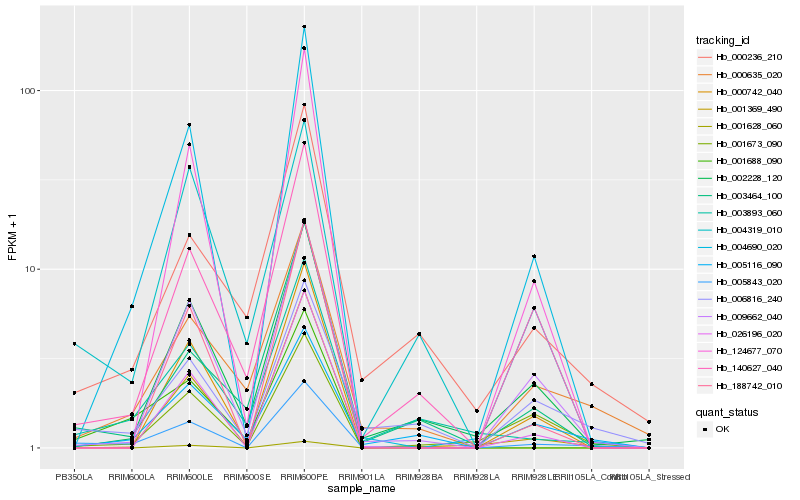
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005843_020 |
0.0 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 2 |
Hb_006816_240 |
0.1699549807 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 3 |
Hb_004690_020 |
0.1718912649 |
- |
- |
PREDICTED: MLP-like protein 328 [Jatropha curcas] |
| 4 |
Hb_009662_040 |
0.1826410906 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 5 |
Hb_005116_090 |
0.1834529956 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003893_060 |
0.1858143599 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000635_020 |
0.1969509441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001369_490 |
0.1986111291 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 33 [Jatropha curcas] |
| 9 |
Hb_001688_090 |
0.2035356416 |
- |
- |
alcohol dehydrogenase 7 [Vitis vinifera] |
| 10 |
Hb_026196_020 |
0.2039289245 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 11 |
Hb_140627_040 |
0.2063244778 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 12 |
Hb_002228_120 |
0.206856654 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 13 |
Hb_004319_010 |
0.2085145128 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 14 |
Hb_001628_060 |
0.2090670991 |
- |
- |
PREDICTED: beta-galactosidase 13-like isoform X2 [Jatropha curcas] |
| 15 |
Hb_003464_100 |
0.2094119809 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g68400 [Jatropha curcas] |
| 16 |
Hb_124677_070 |
0.2104937222 |
- |
- |
PREDICTED: DNA-damage-repair/toleration protein DRT100-like [Jatropha curcas] |
| 17 |
Hb_188742_010 |
0.2108906762 |
- |
- |
hypothetical protein POPTR_0019s12290g [Populus trichocarpa] |
| 18 |
Hb_000742_040 |
0.2109395683 |
- |
- |
PREDICTED: caffeoylshikimate esterase-like [Jatropha curcas] |
| 19 |
Hb_001673_090 |
0.2109983462 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 20 |
Hb_000236_210 |
0.2115048459 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |