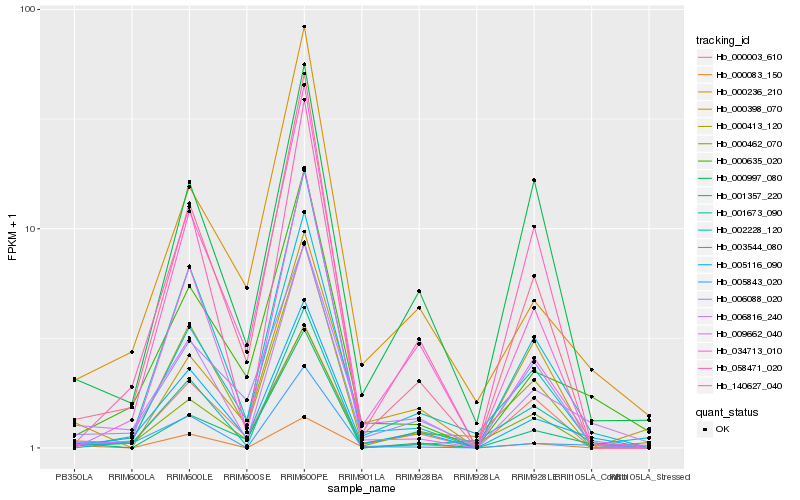
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006816_240 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 2 |
Hb_005116_090 |
0.1203399713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000635_020 |
0.14625975 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000236_210 |
0.1640599176 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 5 |
Hb_005843_020 |
0.1699549807 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 6 |
Hb_003544_080 |
0.1800750805 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 7 |
Hb_000997_080 |
0.1804814248 |
- |
- |
PREDICTED: protein WVD2-like 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006088_020 |
0.1830327665 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 9 |
Hb_002228_120 |
0.1839181666 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 10 |
Hb_140627_040 |
0.1849179282 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 11 |
Hb_000003_610 |
0.1859489136 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase [Jatropha curcas] |
| 12 |
Hb_000083_150 |
0.1867531387 |
- |
- |
PREDICTED: uncharacterized protein LOC105631732 [Jatropha curcas] |
| 13 |
Hb_000462_070 |
0.1872542699 |
- |
- |
PREDICTED: probable pectinesterase 15 [Jatropha curcas] |
| 14 |
Hb_001357_220 |
0.1935334434 |
- |
- |
dopamine beta-monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_001673_090 |
0.1943073214 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |
| 16 |
Hb_000413_120 |
0.1953085666 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 17 |
Hb_009662_040 |
0.1961657271 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 18 |
Hb_058471_020 |
0.1981784275 |
- |
- |
PREDICTED: expansin-B3-like [Jatropha curcas] |
| 19 |
Hb_000398_070 |
0.2005077336 |
- |
- |
hypothetical protein VITISV_011880 [Vitis vinifera] |
| 20 |
Hb_034713_010 |
0.2009080413 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |