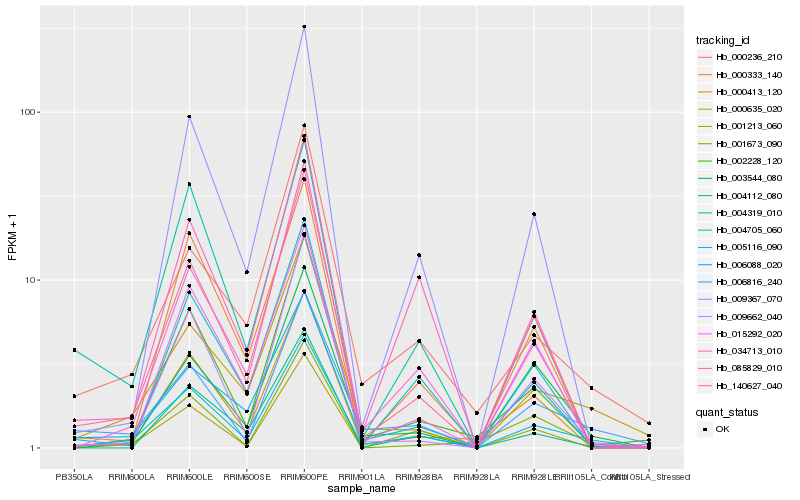
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005116_090 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000635_020 |
0.1200658986 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006816_240 |
0.1203399713 |
- |
- |
PREDICTED: uncharacterized protein LOC105647443 [Jatropha curcas] |
| 4 |
Hb_034713_010 |
0.1366626237 |
- |
- |
PREDICTED: uncharacterized protein LOC105133378 [Populus euphratica] |
| 5 |
Hb_002228_120 |
0.1369095322 |
- |
- |
potassium channel beta, putative [Ricinus communis] |
| 6 |
Hb_140627_040 |
0.1373471208 |
- |
- |
PREDICTED: vesicle-associated protein 4-1-like [Jatropha curcas] |
| 7 |
Hb_015292_020 |
0.1465319753 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_009367_070 |
0.1484970855 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 9 |
Hb_009662_040 |
0.1485450843 |
- |
- |
PREDICTED: uncharacterized protein LOC105114791 [Populus euphratica] |
| 10 |
Hb_004705_060 |
0.1501345189 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 11 |
Hb_003544_080 |
0.1507442921 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 12 |
Hb_000413_120 |
0.1512999453 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g22104-like [Jatropha curcas] |
| 13 |
Hb_000236_210 |
0.1522065982 |
- |
- |
hypothetical protein JCGZ_01083 [Jatropha curcas] |
| 14 |
Hb_006088_020 |
0.1527151307 |
- |
- |
Receptor protein kinase CLAVATA1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000333_140 |
0.1531818831 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 16 |
Hb_001213_060 |
0.1568401884 |
- |
- |
PREDICTED: uncharacterized protein LOC105646662 [Jatropha curcas] |
| 17 |
Hb_004112_080 |
0.1571678986 |
- |
- |
PREDICTED: leucine-rich repeat receptor-like tyrosine-protein kinase At2g41820 [Jatropha curcas] |
| 18 |
Hb_085829_010 |
0.1576325337 |
- |
- |
PREDICTED: homeobox protein 2-like [Jatropha curcas] |
| 19 |
Hb_004319_010 |
0.1601876205 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 20 |
Hb_001673_090 |
0.1652808553 |
- |
- |
hypothetical protein POPTR_0006s21590g [Populus trichocarpa] |