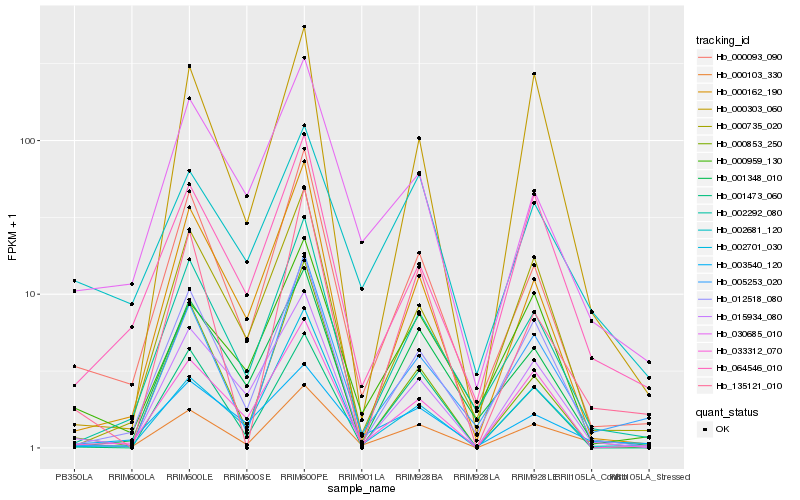
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_330 |
0.0 |
- |
- |
PREDICTED: meiotic recombination protein DMC1 homolog isoform X1 [Glycine max] |
| 2 |
Hb_000093_090 |
0.1380790538 |
- |
- |
PREDICTED: UDP-arabinopyranose mutase 3 [Populus euphratica] |
| 3 |
Hb_015934_080 |
0.1520186788 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12 [Jatropha curcas] |
| 4 |
Hb_012518_080 |
0.1528711037 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 11-like [Jatropha curcas] |
| 5 |
Hb_002292_080 |
0.1539392979 |
- |
- |
transferase, transferring glycosyl groups, putative [Ricinus communis] |
| 6 |
Hb_064546_010 |
0.165297537 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 7 |
Hb_033312_070 |
0.1682443841 |
- |
- |
hypothetical protein POPTR_0017s02960g [Populus trichocarpa] |
| 8 |
Hb_002681_120 |
0.1696210009 |
- |
- |
PREDICTED: GDSL esterase/lipase At5g14450 [Jatropha curcas] |
| 9 |
Hb_005253_020 |
0.1704734564 |
- |
- |
PREDICTED: uncharacterized protein LOC105629998 [Jatropha curcas] |
| 10 |
Hb_030685_010 |
0.1767088571 |
- |
- |
- |
| 11 |
Hb_003540_120 |
0.1776445133 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_000162_190 |
0.1786998896 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 13 |
Hb_000735_020 |
0.1787508454 |
- |
- |
Auxin-induced in root cultures protein 12 precursor, putative [Ricinus communis] |
| 14 |
Hb_135121_010 |
0.1798068897 |
- |
- |
PREDICTED: uncharacterized protein LOC105635544 [Jatropha curcas] |
| 15 |
Hb_000303_060 |
0.1802249626 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 16 |
Hb_000959_130 |
0.1810241122 |
- |
- |
organic anion transporter, putative [Ricinus communis] |
| 17 |
Hb_001348_010 |
0.1819306283 |
- |
- |
PREDICTED: lysM domain-containing GPI-anchored protein 1 [Jatropha curcas] |
| 18 |
Hb_000853_250 |
0.1821558573 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 19 |
Hb_002701_030 |
0.182257532 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 20 |
Hb_001473_060 |
0.1828934795 |
- |
- |
PREDICTED: gibberellin 2-beta-dioxygenase 8 [Jatropha curcas] |