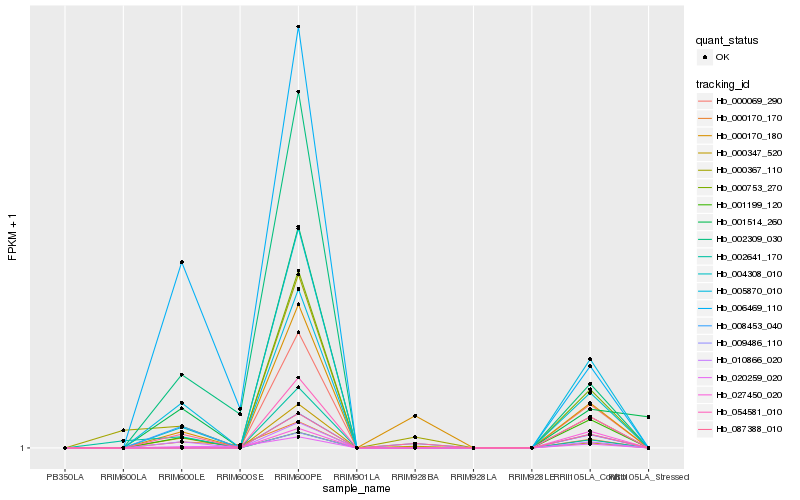
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005870_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_010866_020 |
0.1255582964 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_000170_170 |
0.2010165997 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 4 |
Hb_087388_010 |
0.2107125319 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 5 |
Hb_004308_010 |
0.214856663 |
- |
- |
hypothetical protein CISIN_1g0004672mg, partial [Citrus sinensis] |
| 6 |
Hb_020259_020 |
0.2350964582 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 4 [Jatropha curcas] |
| 7 |
Hb_000347_520 |
0.2470205465 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 8 |
Hb_008453_040 |
0.2554750968 |
- |
- |
PREDICTED: expansin-A11 [Jatropha curcas] |
| 9 |
Hb_054581_010 |
0.2565330794 |
- |
- |
Type I inositol-1,4,5-trisphosphate 5-phosphatase CVP2 [Theobroma cacao] |
| 10 |
Hb_000069_290 |
0.2652697554 |
- |
- |
hypothetical protein POPTR_0014s09500g [Populus trichocarpa] |
| 11 |
Hb_001199_120 |
0.2753753176 |
- |
- |
PREDICTED: uncharacterized protein LOC103437171 [Malus domestica] |
| 12 |
Hb_000170_180 |
0.2804968235 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 13 |
Hb_000753_270 |
0.280608038 |
- |
- |
- |
| 14 |
Hb_027450_020 |
0.2817722398 |
- |
- |
PREDICTED: B3 domain-containing protein At2g31420-like [Citrus sinensis] |
| 15 |
Hb_002641_170 |
0.2824015566 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 16 |
Hb_009486_110 |
0.2825029367 |
- |
- |
PREDICTED: rac-like GTP-binding protein RAC2 [Jatropha curcas] |
| 17 |
Hb_006469_110 |
0.2828710771 |
- |
- |
hypothetical protein JCGZ_12008 [Jatropha curcas] |
| 18 |
Hb_001514_260 |
0.2883282346 |
- |
- |
- |
| 19 |
Hb_002309_030 |
0.2927458908 |
- |
- |
Glycerol dehydrogenase large subunit, putative [Theobroma cacao] |
| 20 |
Hb_000367_110 |
0.2991152755 |
- |
- |
PREDICTED: monodehydroascorbate reductase, chloroplastic isoform X1 [Sesamum indicum] |