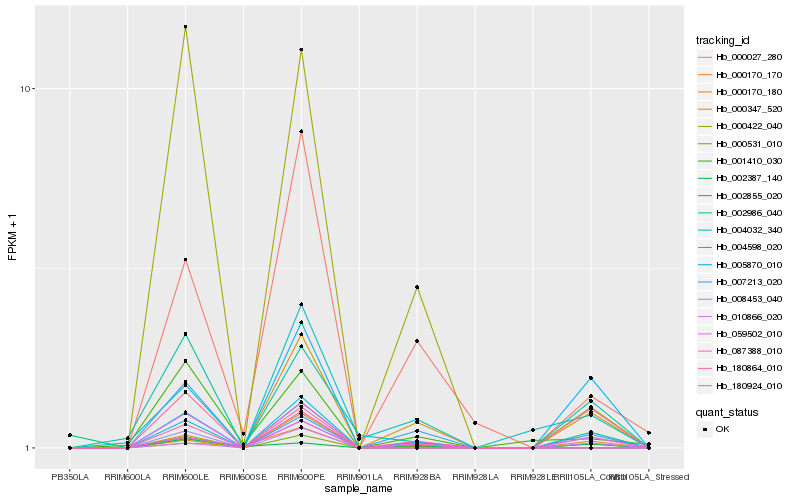
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008453_040 |
0.0 |
- |
- |
PREDICTED: expansin-A11 [Jatropha curcas] |
| 2 |
Hb_087388_010 |
0.0863130391 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 3 |
Hb_000347_520 |
0.232660699 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |
| 4 |
Hb_002986_040 |
0.2348080422 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 5 |
Hb_010866_020 |
0.2365445199 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_180924_010 |
0.248692818 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 7 |
Hb_004032_340 |
0.2512510916 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 8 |
Hb_005870_010 |
0.2554750968 |
- |
- |
- |
| 9 |
Hb_000027_280 |
0.2574436432 |
- |
- |
PREDICTED: uncharacterized protein LOC105642051 [Jatropha curcas] |
| 10 |
Hb_007213_020 |
0.2783070283 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002387_140 |
0.2798408135 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like [Jatropha curcas] |
| 12 |
Hb_000170_180 |
0.2874990992 |
- |
- |
PREDICTED: uncharacterized protein LOC105109433 [Populus euphratica] |
| 13 |
Hb_000422_040 |
0.2928722019 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 14 |
Hb_001410_030 |
0.2962396986 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 15 |
Hb_000170_170 |
0.3003945188 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase 5 [Jatropha curcas] |
| 16 |
Hb_002855_020 |
0.3093405517 |
- |
- |
- |
| 17 |
Hb_004598_020 |
0.3094190443 |
transcription factor |
TF Family: ERF |
Transcriptional factor TINY, putative [Ricinus communis] |
| 18 |
Hb_180864_010 |
0.309428209 |
- |
- |
hypothetical protein B456_010G097000 [Gossypium raimondii] |
| 19 |
Hb_000531_010 |
0.311856125 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0018s14830g [Populus trichocarpa] |
| 20 |
Hb_059502_010 |
0.3120229935 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |