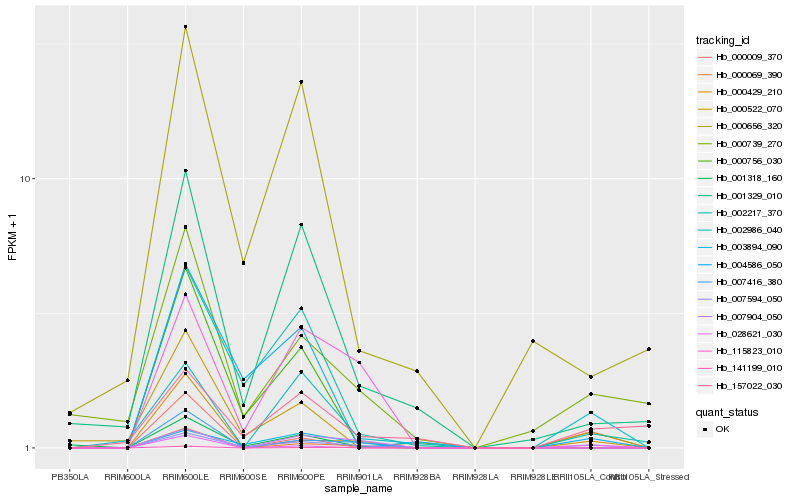
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007594_050 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_002217_370 |
0.2456865428 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105643685 [Jatropha curcas] |
| 3 |
Hb_002986_040 |
0.2590981626 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 4 |
Hb_001329_010 |
0.2642066563 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 5 |
Hb_000739_270 |
0.266832886 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 6 |
Hb_115823_010 |
0.2683013897 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000429_210 |
0.2719541737 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 8 |
Hb_003894_090 |
0.2725458803 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000069_390 |
0.2818993604 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 10 |
Hb_000522_070 |
0.2843787155 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 11 |
Hb_007416_380 |
0.2865719504 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 12 |
Hb_028621_030 |
0.2898504629 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 13 |
Hb_000656_320 |
0.2913582677 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 14 |
Hb_007904_050 |
0.2952892622 |
- |
- |
hypothetical protein JCGZ_17883 [Jatropha curcas] |
| 15 |
Hb_000009_370 |
0.2964378072 |
- |
- |
hypothetical protein JCGZ_12284 [Jatropha curcas] |
| 16 |
Hb_157022_030 |
0.2966311467 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 17 |
Hb_000756_030 |
0.2969411443 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_001318_160 |
0.297860749 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 19 |
Hb_141199_010 |
0.2980690791 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_004586_050 |
0.2999343732 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |