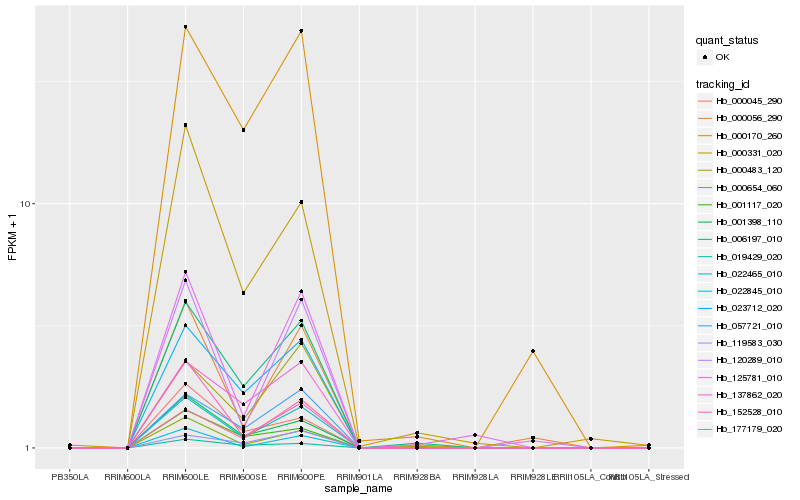
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022465_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_06012 [Jatropha curcas] |
| 2 |
Hb_023712_020 |
0.0777005118 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20669 [Jatropha curcas] |
| 3 |
Hb_006197_010 |
0.0838202296 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 4 |
Hb_057721_010 |
0.088326371 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 5 |
Hb_000654_060 |
0.0885233798 |
- |
- |
hypothetical protein CICLE_v10027550mg [Citrus clementina] |
| 6 |
Hb_001398_110 |
0.106993924 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 7 |
Hb_177179_020 |
0.108315181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_137862_020 |
0.1146071838 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 9 |
Hb_022845_010 |
0.1153551002 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 10 |
Hb_000483_120 |
0.1198089955 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0004s12530g [Populus trichocarpa] |
| 11 |
Hb_001117_020 |
0.1250923589 |
- |
- |
hypothetical protein CICLE_v10008579mg [Citrus clementina] |
| 12 |
Hb_000331_020 |
0.1251068724 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_125781_010 |
0.1261408 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 14 |
Hb_019429_020 |
0.1265646254 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 15 |
Hb_119583_030 |
0.1295045855 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 16 |
Hb_000170_260 |
0.130315063 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 17 |
Hb_152528_010 |
0.1316097328 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 18 |
Hb_120289_010 |
0.1325518345 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 19 |
Hb_000056_290 |
0.1360456428 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000045_290 |
0.1376609749 |
- |
- |
- |