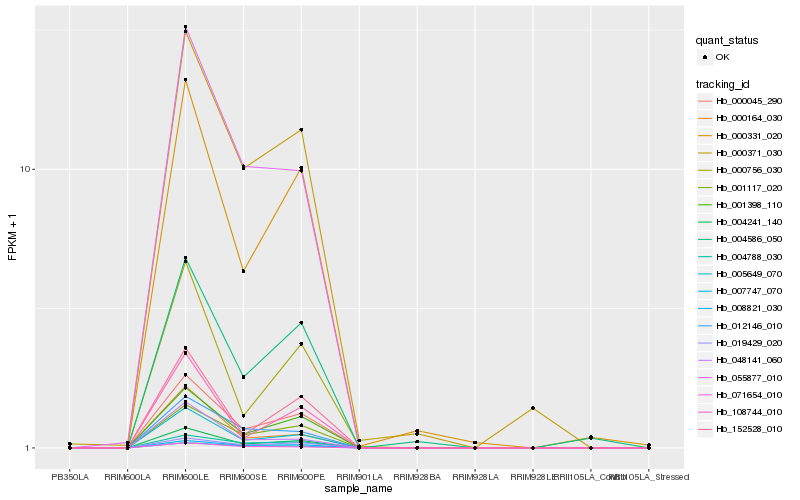
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004241_140 |
0.0 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 2 |
Hb_000045_290 |
0.0247995343 |
- |
- |
- |
| 3 |
Hb_005649_070 |
0.0285907808 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001398_110 |
0.0514075735 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 5 |
Hb_001117_020 |
0.0702950778 |
- |
- |
hypothetical protein CICLE_v10008579mg [Citrus clementina] |
| 6 |
Hb_055877_010 |
0.0892402047 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_019429_020 |
0.0893559867 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 8 |
Hb_000331_020 |
0.0899595466 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 9 |
Hb_008821_030 |
0.0919874878 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 10 |
Hb_152528_010 |
0.0966664945 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 11 |
Hb_000756_030 |
0.0998788987 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_012146_010 |
0.1050895991 |
- |
- |
PREDICTED: uncharacterized protein LOC105111251 [Populus euphratica] |
| 13 |
Hb_108744_010 |
0.1066834781 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 14 |
Hb_000371_030 |
0.1078061763 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: anthocyanidin 3-O-glucosyltransferase 5 [Prunus mume] |
| 15 |
Hb_071654_010 |
0.1084345759 |
- |
- |
non-ltr retroelement reverse transcriptase [Rosa rugosa] |
| 16 |
Hb_004788_030 |
0.1087319457 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_007747_070 |
0.108734799 |
- |
- |
PREDICTED: casparian strip membrane protein 1-like [Jatropha curcas] |
| 18 |
Hb_000164_030 |
0.1105309848 |
- |
- |
PREDICTED: RING-H2 finger protein ATL14-like [Zea mays] |
| 19 |
Hb_048141_060 |
0.117997721 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein ZAT3 [Populus euphratica] |
| 20 |
Hb_004586_050 |
0.1181741538 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |