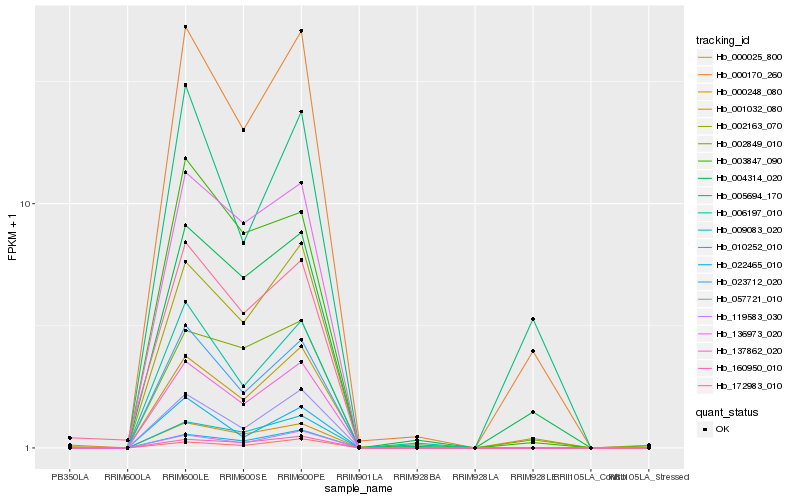
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000170_260 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_001032_080 |
0.0734413817 |
- |
- |
Ankyrin repeat protein [Medicago truncatula] |
| 3 |
Hb_137862_020 |
0.0852052039 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 4 |
Hb_004314_020 |
0.0867252453 |
- |
- |
Auxin-responsive GH3 family protein [Theobroma cacao] |
| 5 |
Hb_023712_020 |
0.0906911679 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20669 [Jatropha curcas] |
| 6 |
Hb_057721_010 |
0.0976831587 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 7 |
Hb_002163_070 |
0.098441601 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 8 |
Hb_000248_080 |
0.1014037001 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 9 |
Hb_006197_010 |
0.1068189184 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 10 |
Hb_010252_010 |
0.1083996667 |
- |
- |
- |
| 11 |
Hb_009083_020 |
0.1106583888 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 12 |
Hb_119583_030 |
0.1113746407 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_005694_170 |
0.1135612736 |
- |
- |
PREDICTED: uncharacterized protein At3g49140 isoform X2 [Jatropha curcas] |
| 14 |
Hb_136973_020 |
0.119683808 |
- |
- |
- |
| 15 |
Hb_172983_010 |
0.1228954018 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_160950_010 |
0.1235425609 |
- |
- |
PREDICTED: uncharacterized protein LOC105761043 [Gossypium raimondii] |
| 17 |
Hb_002849_010 |
0.1240776731 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |
| 18 |
Hb_000025_800 |
0.1299360738 |
- |
- |
hypothetical protein VITISV_020143 [Vitis vinifera] |
| 19 |
Hb_022465_010 |
0.130315063 |
- |
- |
hypothetical protein JCGZ_06012 [Jatropha curcas] |
| 20 |
Hb_003847_090 |
0.1345344898 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |