| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
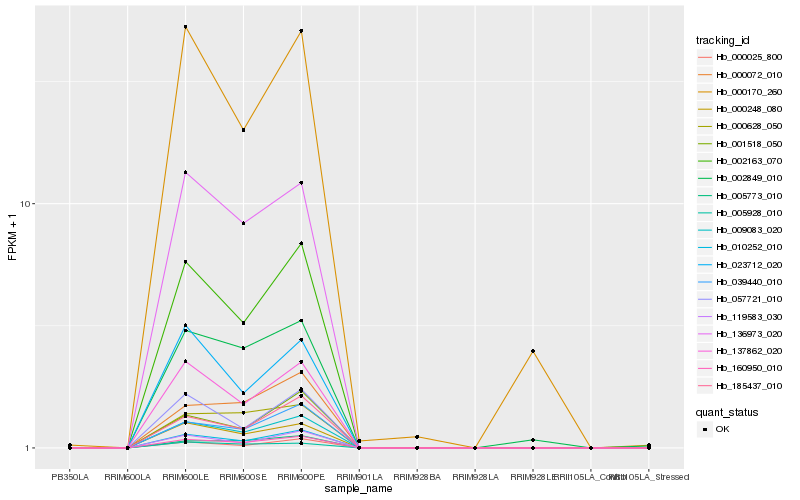
Hb_009083_020 |
0.0 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 2 |
Hb_160950_010 |
0.018990972 |
- |
- |
PREDICTED: uncharacterized protein LOC105761043 [Gossypium raimondii] |
| 3 |
Hb_002163_070 |
0.0293248235 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 4 |
Hb_010252_010 |
0.0303780635 |
- |
- |
- |
| 5 |
Hb_000248_080 |
0.0537375067 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 6 |
Hb_137862_020 |
0.0588866356 |
- |
- |
Leucine-rich repeat receptor protein kinase EXS precursor, putative [Ricinus communis] |
| 7 |
Hb_039440_010 |
0.0705117471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_136973_020 |
0.0750294068 |
- |
- |
- |
| 9 |
Hb_119583_030 |
0.0758164201 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 10 |
Hb_185437_010 |
0.0780040545 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 11 |
Hb_000025_800 |
0.0850889681 |
- |
- |
hypothetical protein VITISV_020143 [Vitis vinifera] |
| 12 |
Hb_057721_010 |
0.0923357064 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 13 |
Hb_005773_010 |
0.0940553556 |
- |
- |
- |
| 14 |
Hb_001518_050 |
0.0958771668 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000628_050 |
0.1053168437 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_023712_020 |
0.1053799277 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20669 [Jatropha curcas] |
| 17 |
Hb_005928_010 |
0.1064876055 |
- |
- |
PREDICTED: phospholipase A1-Igamma1, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_000072_010 |
0.1091286312 |
- |
- |
hypothetical protein B456_006G246700 [Gossypium raimondii] |
| 19 |
Hb_000170_260 |
0.1106583888 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 20 |
Hb_002849_010 |
0.1127031653 |
- |
- |
PREDICTED: putative UPF0481 protein At3g02645 [Jatropha curcas] |