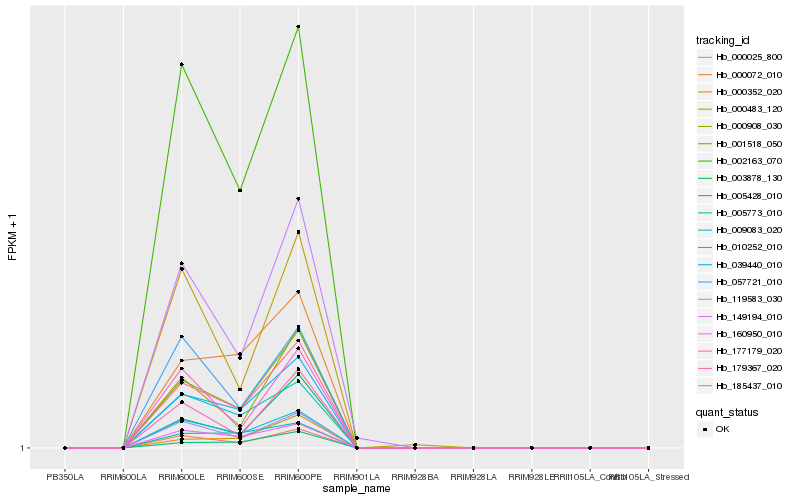
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001518_050 |
0.0 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 2 |
Hb_185437_010 |
0.0182593472 |
- |
- |
PREDICTED: uncharacterized protein LOC105637003 [Jatropha curcas] |
| 3 |
Hb_039440_010 |
0.046213348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000025_800 |
0.0477323838 |
- |
- |
hypothetical protein VITISV_020143 [Vitis vinifera] |
| 5 |
Hb_119583_030 |
0.0727627922 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_010252_010 |
0.0738460646 |
- |
- |
- |
| 7 |
Hb_160950_010 |
0.0879195756 |
- |
- |
PREDICTED: uncharacterized protein LOC105761043 [Gossypium raimondii] |
| 8 |
Hb_000908_030 |
0.0890902369 |
- |
- |
small heat-shock protein, putative [Ricinus communis] |
| 9 |
Hb_003878_130 |
0.0897569173 |
- |
- |
- |
| 10 |
Hb_149194_010 |
0.0905240578 |
- |
- |
hypothetical protein JCGZ_17021 [Jatropha curcas] |
| 11 |
Hb_002163_070 |
0.0911329791 |
- |
- |
hypothetical protein JCGZ_13728 [Jatropha curcas] |
| 12 |
Hb_009083_020 |
0.0958771668 |
- |
- |
PREDICTED: thaumatin-like protein 1b [Jatropha curcas] |
| 13 |
Hb_179367_020 |
0.1013629578 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 14 |
Hb_005428_010 |
0.1027286054 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_000072_010 |
0.111568001 |
- |
- |
hypothetical protein B456_006G246700 [Gossypium raimondii] |
| 16 |
Hb_057721_010 |
0.1147688215 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |
| 17 |
Hb_000483_120 |
0.1172251005 |
transcription factor |
TF Family: MYB |
hypothetical protein POPTR_0004s12530g [Populus trichocarpa] |
| 18 |
Hb_177179_020 |
0.1221721582 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_005773_010 |
0.1236223607 |
- |
- |
- |
| 20 |
Hb_000352_020 |
0.1270148665 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor LEP [Jatropha curcas] |