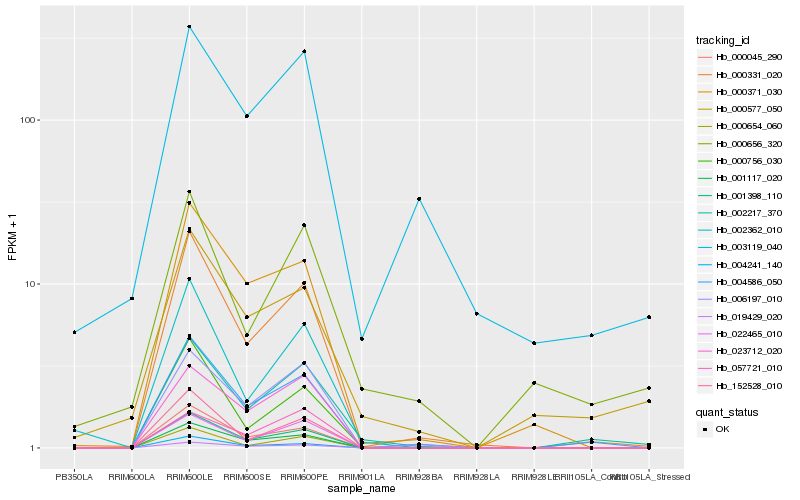
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002217_370 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105643685 [Jatropha curcas] |
| 2 |
Hb_004586_050 |
0.1291922739 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH30-like [Jatropha curcas] |
| 3 |
Hb_000331_020 |
0.135933163 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_006197_010 |
0.1605320703 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 5 |
Hb_022465_010 |
0.1610857437 |
- |
- |
hypothetical protein JCGZ_06012 [Jatropha curcas] |
| 6 |
Hb_001398_110 |
0.162574152 |
- |
- |
Early nodulin 55-2 precursor, putative [Ricinus communis] |
| 7 |
Hb_000656_320 |
0.1700582358 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 3-like [Jatropha curcas] |
| 8 |
Hb_023712_020 |
0.1703047913 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_20669 [Jatropha curcas] |
| 9 |
Hb_001117_020 |
0.1703163785 |
- |
- |
hypothetical protein CICLE_v10008579mg [Citrus clementina] |
| 10 |
Hb_000654_060 |
0.170950953 |
- |
- |
hypothetical protein CICLE_v10027550mg [Citrus clementina] |
| 11 |
Hb_019429_020 |
0.1728080155 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000577_050 |
0.1738480954 |
- |
- |
- |
| 13 |
Hb_000045_290 |
0.1750964953 |
- |
- |
- |
| 14 |
Hb_002362_010 |
0.179641442 |
- |
- |
hypothetical protein JCGZ_12328 [Jatropha curcas] |
| 15 |
Hb_000756_030 |
0.1842392689 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_152528_010 |
0.184966749 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 17 |
Hb_004241_140 |
0.1860862156 |
- |
- |
S-locus lectin protein kinase family protein, putative [Theobroma cacao] |
| 18 |
Hb_000371_030 |
0.186318746 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: anthocyanidin 3-O-glucosyltransferase 5 [Prunus mume] |
| 19 |
Hb_003119_040 |
0.1882125384 |
- |
- |
PREDICTED: classical arabinogalactan protein 1-like [Populus euphratica] |
| 20 |
Hb_057721_010 |
0.1934684363 |
- |
- |
hypothetical protein JCGZ_26658 [Jatropha curcas] |