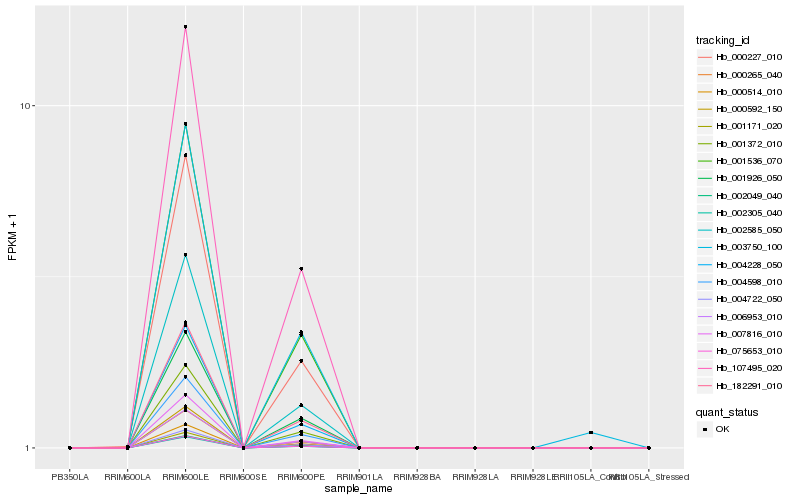
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003750_100 |
0.0 |
- |
- |
HMGI/Y family protein [Populus trichocarpa] |
| 2 |
Hb_002049_040 |
0.0777275058 |
- |
- |
PREDICTED: protein UNUSUAL FLORAL ORGANS [Jatropha curcas] |
| 3 |
Hb_006953_010 |
0.0777375924 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_182291_010 |
0.0777439387 |
- |
- |
hypothetical protein CICLE_v10029654mg [Citrus clementina] |
| 5 |
Hb_004598_010 |
0.0777806391 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase-like 5 [Jatropha curcas] |
| 6 |
Hb_001372_010 |
0.0777937177 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_107495_020 |
0.0778387655 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 8 |
Hb_001536_070 |
0.0778823725 |
- |
- |
hypothetical protein JCGZ_20053 [Jatropha curcas] |
| 9 |
Hb_000265_040 |
0.0778897674 |
- |
- |
PREDICTED: subtilisin-like protease, partial [Prunus mume] |
| 10 |
Hb_001171_020 |
0.0779796222 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 3-like [Citrus sinensis] |
| 11 |
Hb_075653_010 |
0.0782519397 |
- |
- |
PREDICTED: tyrosine decarboxylase 1-like [Jatropha curcas] |
| 12 |
Hb_002305_040 |
0.0783704776 |
- |
- |
TMV resistance protein N, putative [Ricinus communis] |
| 13 |
Hb_000514_010 |
0.078599481 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 14 |
Hb_004228_050 |
0.0792799069 |
- |
- |
PREDICTED: tubulin beta chain-like [Jatropha curcas] |
| 15 |
Hb_000592_150 |
0.0794686813 |
- |
- |
Os05g0413200 [Oryza sativa Japonica Group] |
| 16 |
Hb_004722_050 |
0.0809030536 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 17 |
Hb_002585_050 |
0.0818182119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001926_050 |
0.0841901747 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 19 |
Hb_000227_010 |
0.0844454413 |
- |
- |
hypothetical protein JCGZ_02114 [Jatropha curcas] |
| 20 |
Hb_007816_010 |
0.0845428026 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |