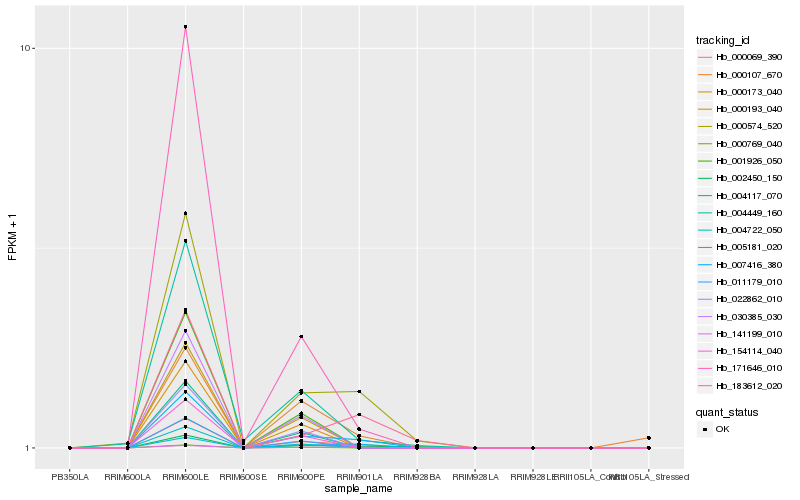
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_390 |
0.0 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 2 |
Hb_007416_380 |
0.0295003856 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 3 |
Hb_000574_520 |
0.1214222537 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 4 |
Hb_141199_010 |
0.1366540704 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_005181_020 |
0.1728980012 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_004449_160 |
0.1785740053 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 7 |
Hb_002450_150 |
0.1871774373 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_171646_010 |
0.1915196393 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 9 |
Hb_000107_670 |
0.1963480053 |
transcription factor |
TF Family: MIKC |
floral homeotic protein AGAMOUS [Jatropha curcas] |
| 10 |
Hb_004117_070 |
0.2022243328 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 11 |
Hb_030385_030 |
0.2023205678 |
- |
- |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 12 |
Hb_011179_010 |
0.2024745076 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 13 |
Hb_001926_050 |
0.2025472932 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 14 |
Hb_000173_040 |
0.2028799971 |
- |
- |
putative xylanase inhibitor, partial [Triticum aestivum] |
| 15 |
Hb_154114_040 |
0.2029386913 |
- |
- |
hypothetical protein B456_001G056600 [Gossypium raimondii] |
| 16 |
Hb_000193_040 |
0.203172232 |
- |
- |
PREDICTED: GDSL esterase/lipase EXL3-like [Jatropha curcas] |
| 17 |
Hb_183612_020 |
0.2032172968 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 18 |
Hb_004722_050 |
0.2033681521 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 19 |
Hb_022862_010 |
0.2033786914 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 20 |
Hb_000769_040 |
0.2038148394 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |