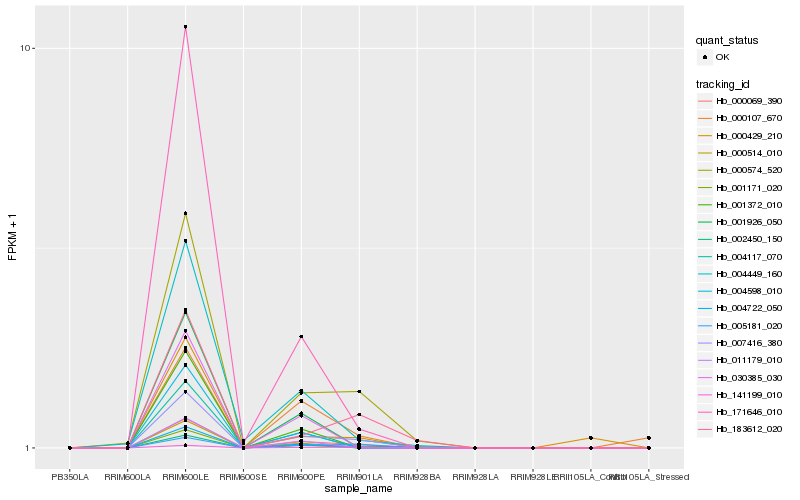
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007416_380 |
0.0 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 2 |
Hb_000069_390 |
0.0295003856 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 3 |
Hb_000574_520 |
0.1089357732 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 4 |
Hb_141199_010 |
0.1136368976 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_005181_020 |
0.1518307572 |
- |
- |
Pectinesterase-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_183612_020 |
0.1842598181 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 7 |
Hb_002450_150 |
0.1851248101 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 8 |
Hb_004449_160 |
0.1955556663 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_171646_010 |
0.2035429396 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 10 |
Hb_000429_210 |
0.2084553994 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 11 |
Hb_000107_670 |
0.2092779036 |
transcription factor |
TF Family: MIKC |
floral homeotic protein AGAMOUS [Jatropha curcas] |
| 12 |
Hb_001926_050 |
0.2238916808 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 13 |
Hb_004722_050 |
0.224010656 |
- |
- |
hypothetical protein POPTR_0011s02010g [Populus trichocarpa] |
| 14 |
Hb_004117_070 |
0.2241011542 |
- |
- |
PREDICTED: GDSL esterase/lipase At1g71691 [Jatropha curcas] |
| 15 |
Hb_000514_010 |
0.2246137021 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 16 |
Hb_030385_030 |
0.2249457922 |
- |
- |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001171_020 |
0.2251004449 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 3-like [Citrus sinensis] |
| 18 |
Hb_011179_010 |
0.2252936537 |
transcription factor |
TF Family: ERF |
hypothetical protein JCGZ_14625 [Jatropha curcas] |
| 19 |
Hb_001372_010 |
0.2254268131 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_004598_010 |
0.2254643774 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase-like 5 [Jatropha curcas] |