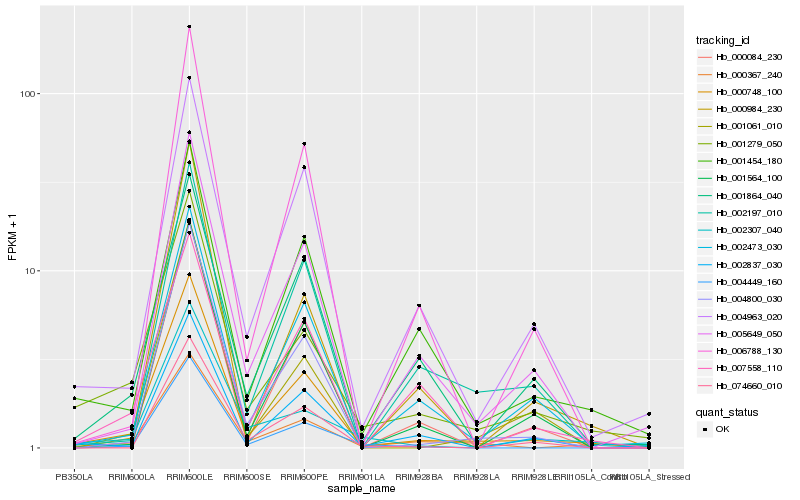
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000748_100 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 2 |
Hb_004800_030 |
0.1123126879 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 3 |
Hb_001279_050 |
0.1209098598 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 4 |
Hb_002473_030 |
0.1302380012 |
- |
- |
PREDICTED: osmotic avoidance abnormal protein 3 [Jatropha curcas] |
| 5 |
Hb_005649_050 |
0.1331775857 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001454_180 |
0.1363685674 |
- |
- |
PREDICTED: uncharacterized protein LOC105643432 [Jatropha curcas] |
| 7 |
Hb_004963_020 |
0.1366561701 |
- |
- |
PREDICTED: patellin-4 [Populus euphratica] |
| 8 |
Hb_000367_240 |
0.1371660519 |
- |
- |
hypothetical protein JCGZ_11682 [Jatropha curcas] |
| 9 |
Hb_006788_130 |
0.1379378879 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 10 |
Hb_001564_100 |
0.1386974818 |
- |
- |
Chromosome-associated kinesin KIF4A, putative [Ricinus communis] |
| 11 |
Hb_002837_030 |
0.1420742325 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_004449_160 |
0.1435783895 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 13 |
Hb_001061_010 |
0.1456978829 |
- |
- |
PREDICTED: uncharacterized protein LOC100243595 [Vitis vinifera] |
| 14 |
Hb_000084_230 |
0.148893312 |
- |
- |
hypothetical protein POPTR_0016s08030g [Populus trichocarpa] |
| 15 |
Hb_000984_230 |
0.1497963437 |
- |
- |
- |
| 16 |
Hb_074660_010 |
0.1503479863 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 17 |
Hb_002197_010 |
0.150376472 |
- |
- |
cyclin d, putative [Ricinus communis] |
| 18 |
Hb_001864_040 |
0.1560273357 |
- |
- |
PREDICTED: kinesin-1 [Jatropha curcas] |
| 19 |
Hb_007558_110 |
0.1561164813 |
- |
- |
PREDICTED: uncharacterized protein LOC105649093 [Jatropha curcas] |
| 20 |
Hb_002307_040 |
0.1586913373 |
- |
- |
DNA binding protein, putative [Ricinus communis] |