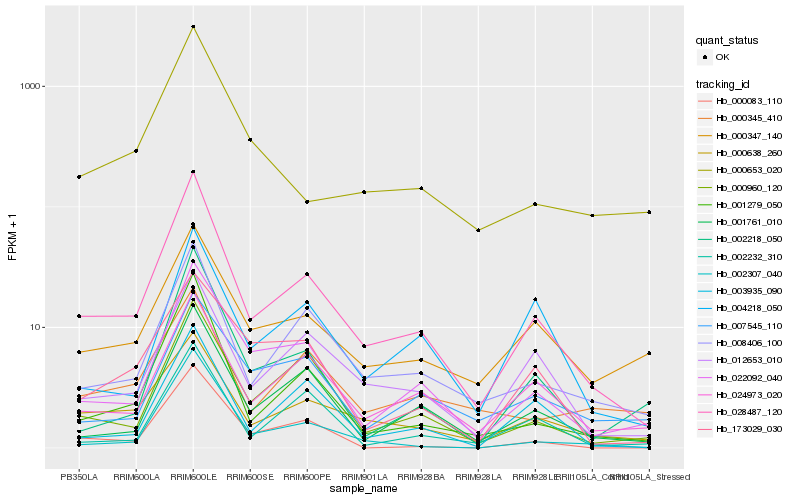
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028487_120 |
0.0 |
- |
- |
PREDICTED: HMG-Y-related protein B-like [Jatropha curcas] |
| 2 |
Hb_001279_050 |
0.119922848 |
- |
- |
PREDICTED: UPF0392 protein RCOM_0530710 [Jatropha curcas] |
| 3 |
Hb_008406_100 |
0.1393301549 |
- |
- |
PREDICTED: protein TPX2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_022092_040 |
0.1436032682 |
- |
- |
PREDICTED: serine/arginine repetitive matrix protein 1 [Jatropha curcas] |
| 5 |
Hb_000638_260 |
0.1437153851 |
- |
- |
hypothetical protein JCGZ_06730 [Jatropha curcas] |
| 6 |
Hb_000960_120 |
0.1529097318 |
- |
- |
PREDICTED: cell division cycle 20.1, cofactor of APC complex-like [Jatropha curcas] |
| 7 |
Hb_024973_020 |
0.1534799589 |
- |
- |
Protein regulator of cytokinesis, putative [Ricinus communis] |
| 8 |
Hb_001761_010 |
0.1572975804 |
- |
- |
PREDICTED: uncharacterized protein LOC105639282 [Jatropha curcas] |
| 9 |
Hb_012653_010 |
0.1641469504 |
- |
- |
PREDICTED: subtilisin-like protease SBT2.5 [Jatropha curcas] |
| 10 |
Hb_002307_040 |
0.1693861857 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_003935_090 |
0.1744272907 |
- |
- |
PREDICTED: uncharacterized protein At4g38062 [Jatropha curcas] |
| 12 |
Hb_000083_110 |
0.1754557541 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 13 |
Hb_002218_050 |
0.1760060754 |
- |
- |
SSXT family protein isoform 2, partial [Theobroma cacao] |
| 14 |
Hb_000347_140 |
0.1872354016 |
- |
- |
PREDICTED: protein FLUORESCENT IN BLUE LIGHT, chloroplastic isoform X2 [Jatropha curcas] |
| 15 |
Hb_000653_020 |
0.1873739991 |
- |
- |
PREDICTED: protein EARLY RESPONSIVE TO DEHYDRATION 15-like [Jatropha curcas] |
| 16 |
Hb_002232_310 |
0.1912361375 |
- |
- |
PREDICTED: uncharacterized protein LOC105635987 [Jatropha curcas] |
| 17 |
Hb_173029_030 |
0.1962264848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000345_410 |
0.1972222905 |
- |
- |
PREDICTED: kinesin-like protein KIFC3 [Jatropha curcas] |
| 19 |
Hb_004218_050 |
0.2004817977 |
- |
- |
annexin [Manihot esculenta] |
| 20 |
Hb_007545_110 |
0.2023443201 |
- |
- |
catalytic, putative [Ricinus communis] |