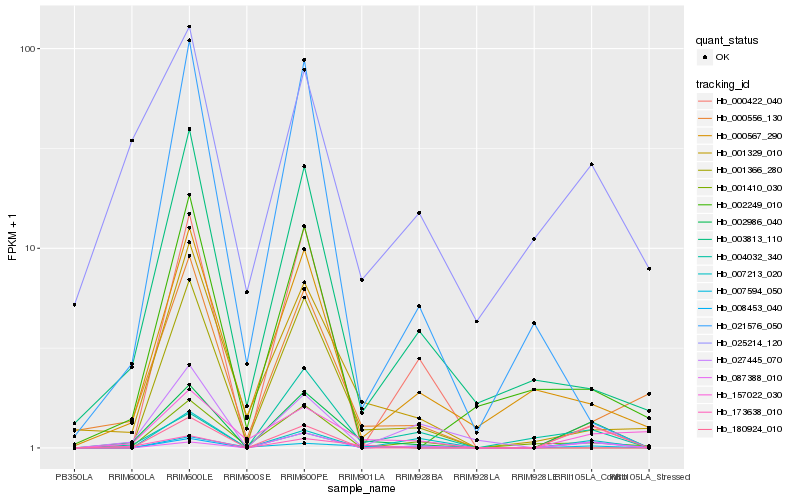
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002986_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639694 [Jatropha curcas] |
| 2 |
Hb_008453_040 |
0.2348080422 |
- |
- |
PREDICTED: expansin-A11 [Jatropha curcas] |
| 3 |
Hb_180924_010 |
0.2385126166 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 4 |
Hb_007594_050 |
0.2590981626 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 5 |
Hb_000556_130 |
0.2785947383 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-3 [Jatropha curcas] |
| 6 |
Hb_001410_030 |
0.2801427333 |
- |
- |
hypothetical protein JCGZ_20419 [Jatropha curcas] |
| 7 |
Hb_001329_010 |
0.2845392038 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 8 |
Hb_007213_020 |
0.2857076691 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003813_110 |
0.2865056427 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_157022_030 |
0.2871286722 |
transcription factor |
TF Family: NF-YB |
ccaat-binding transcription factor subunit A, putative [Ricinus communis] |
| 11 |
Hb_000567_290 |
0.2888584841 |
- |
- |
hypothetical protein RCOM_0303940 [Ricinus communis] |
| 12 |
Hb_087388_010 |
0.2936234276 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor DYT1 [Jatropha curcas] |
| 13 |
Hb_025214_120 |
0.2956352184 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 13 [Jatropha curcas] |
| 14 |
Hb_002249_010 |
0.2999307855 |
- |
- |
serine carboxypeptidase, putative [Ricinus communis] |
| 15 |
Hb_000422_040 |
0.3040473825 |
- |
- |
hypothetical protein CICLE_v10013147mg [Citrus clementina] |
| 16 |
Hb_001366_280 |
0.3050213307 |
- |
- |
PREDICTED: uncharacterized protein LOC105642732 [Jatropha curcas] |
| 17 |
Hb_027445_070 |
0.3080206217 |
- |
- |
PREDICTED: denticleless protein homolog [Jatropha curcas] |
| 18 |
Hb_004032_340 |
0.3097405806 |
- |
- |
PREDICTED: mitotic spindle checkpoint protein BUBR1 [Jatropha curcas] |
| 19 |
Hb_173638_010 |
0.3097466963 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_021576_050 |
0.3104632181 |
- |
- |
PREDICTED: early nodulin-like protein 1 [Jatropha curcas] |