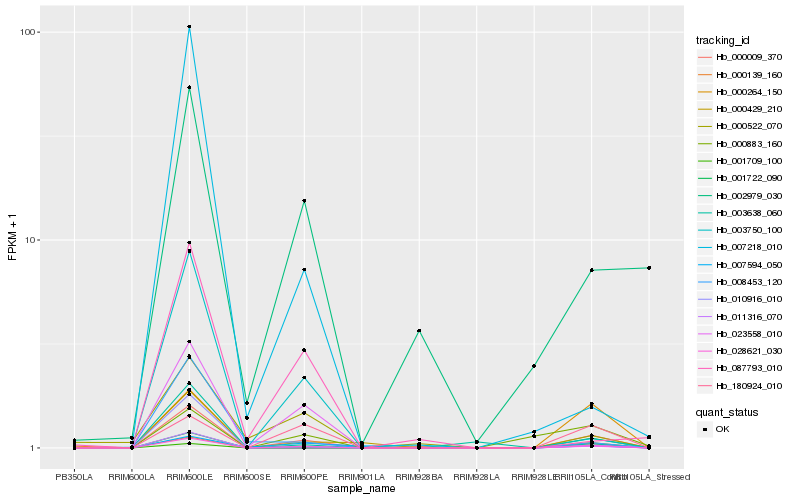
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000009_370 |
0.0 |
- |
- |
hypothetical protein JCGZ_12284 [Jatropha curcas] |
| 2 |
Hb_028621_030 |
0.0146748029 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 3 |
Hb_000429_210 |
0.2430065286 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 4 |
Hb_011316_070 |
0.2437222965 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_003750_100 |
0.2602005709 |
- |
- |
HMGI/Y family protein [Populus trichocarpa] |
| 6 |
Hb_180924_010 |
0.2632479921 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 7 |
Hb_008453_120 |
0.2840890923 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0005s26480g [Populus trichocarpa] |
| 8 |
Hb_001709_100 |
0.2858778046 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like [Jatropha curcas] |
| 9 |
Hb_001722_090 |
0.2863406655 |
- |
- |
cyanohydrin UDP-glucosyltransferase UGT85K5 [Manihot esculenta] |
| 10 |
Hb_010916_010 |
0.2884946191 |
- |
- |
terpene synthase [Populus trichocarpa] |
| 11 |
Hb_003638_060 |
0.2915489965 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 12 |
Hb_000522_070 |
0.2917445018 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 13 |
Hb_023558_010 |
0.2927889014 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 14 |
Hb_000139_160 |
0.2940514145 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_007594_050 |
0.2964378072 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002979_030 |
0.3015993352 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000264_150 |
0.3037821008 |
- |
- |
PREDICTED: uncharacterized protein LOC103870759 [Brassica rapa] |
| 18 |
Hb_000883_160 |
0.3050510466 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_087793_010 |
0.3061304886 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007218_010 |
0.3069739117 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |