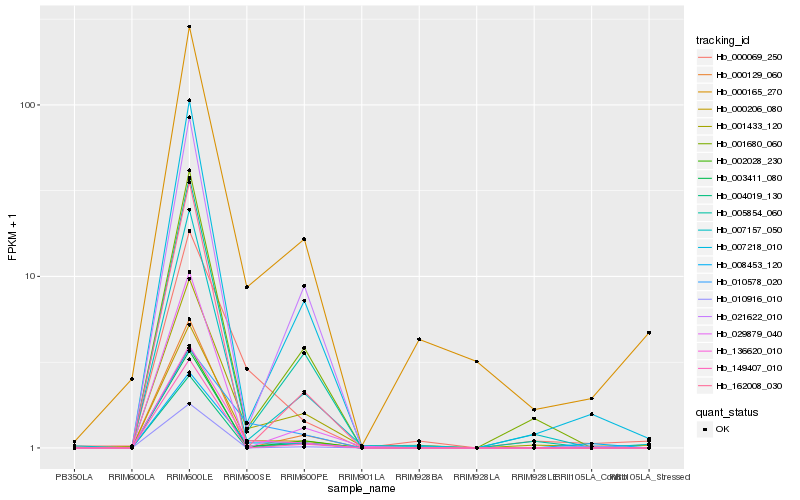
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008453_120 |
0.0 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0005s26480g [Populus trichocarpa] |
| 2 |
Hb_136620_010 |
0.1297039792 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 10-like [Jatropha curcas] |
| 3 |
Hb_000206_080 |
0.134376251 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein Myb4-like [Jatropha curcas] |
| 4 |
Hb_001433_120 |
0.1463682973 |
- |
- |
hypothetical protein CICLE_v10003574mg, partial [Citrus clementina] |
| 5 |
Hb_007218_010 |
0.1496036949 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 6 |
Hb_003411_080 |
0.154817178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_010916_010 |
0.1571186089 |
- |
- |
terpene synthase [Populus trichocarpa] |
| 8 |
Hb_002028_230 |
0.1678349169 |
- |
- |
Pectinesterase-3 precursor, putative [Ricinus communis] |
| 9 |
Hb_005854_060 |
0.1695239233 |
- |
- |
short-chain dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000069_250 |
0.1704662024 |
- |
- |
hypothetical protein JCGZ_14250 [Jatropha curcas] |
| 11 |
Hb_000165_270 |
0.1715255393 |
transcription factor |
TF Family: Tify |
JAZ10 [Hevea brasiliensis] |
| 12 |
Hb_001680_060 |
0.1797444568 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_007157_050 |
0.1825199174 |
- |
- |
hypothetical protein RCOM_0711500 [Ricinus communis] |
| 14 |
Hb_000129_060 |
0.1851996744 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2-like isoform X1 [Populus euphratica] |
| 15 |
Hb_162008_030 |
0.1854406498 |
- |
- |
cytochrome p450 [Ageratina adenophora] |
| 16 |
Hb_004019_130 |
0.1854576252 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 6 [Jatropha curcas] |
| 17 |
Hb_021622_010 |
0.1877461216 |
- |
- |
hypothetical protein POPTR_0002s06160g [Populus trichocarpa] |
| 18 |
Hb_149407_010 |
0.1884094637 |
- |
- |
PREDICTED: uncharacterized protein LOC105643999 [Jatropha curcas] |
| 19 |
Hb_029879_040 |
0.1884619014 |
- |
- |
PREDICTED: cytochrome P450 77A4-like [Jatropha curcas] |
| 20 |
Hb_010578_020 |
0.1885893917 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |