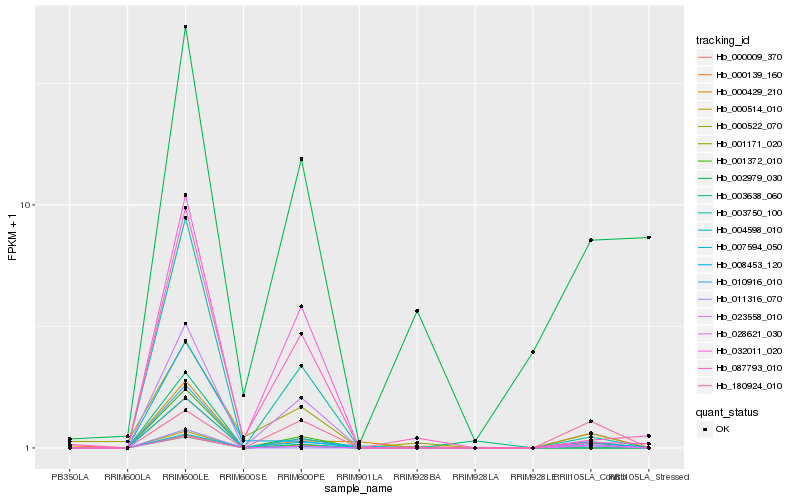
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_028621_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 2 |
Hb_000009_370 |
0.0146748029 |
- |
- |
hypothetical protein JCGZ_12284 [Jatropha curcas] |
| 3 |
Hb_000429_210 |
0.2383519872 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 4 |
Hb_011316_070 |
0.2490284391 |
- |
- |
acyltransferase, putative [Ricinus communis] |
| 5 |
Hb_003750_100 |
0.2490961504 |
- |
- |
HMGI/Y family protein [Populus trichocarpa] |
| 6 |
Hb_180924_010 |
0.2626910088 |
- |
- |
CPI-4 [Morus alba var. atropurpurea] |
| 7 |
Hb_008453_120 |
0.2799409409 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0005s26480g [Populus trichocarpa] |
| 8 |
Hb_023558_010 |
0.2801077631 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 9 |
Hb_000522_070 |
0.2837835064 |
- |
- |
PREDICTED: uncharacterized protein LOC105632109 [Jatropha curcas] |
| 10 |
Hb_010916_010 |
0.2852027414 |
- |
- |
terpene synthase [Populus trichocarpa] |
| 11 |
Hb_003638_060 |
0.2884708404 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-like [Jatropha curcas] |
| 12 |
Hb_007594_050 |
0.2898504629 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 13 |
Hb_087793_010 |
0.2950417997 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002979_030 |
0.2953076763 |
- |
- |
PREDICTED: serine/threonine-protein kinase Aurora-1 isoform X2 [Jatropha curcas] |
| 15 |
Hb_032011_020 |
0.2969381741 |
- |
- |
alpha-farnesene synthase [Ricinus communis] |
| 16 |
Hb_000139_160 |
0.2974300724 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001171_020 |
0.2974496951 |
- |
- |
PREDICTED: gibberellin 3-beta-dioxygenase 3-like [Citrus sinensis] |
| 18 |
Hb_000514_010 |
0.2974736942 |
- |
- |
PREDICTED: probable indole-3-pyruvate monooxygenase YUCCA4 [Jatropha curcas] |
| 19 |
Hb_001372_010 |
0.2974754007 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 20 |
Hb_004598_010 |
0.2974798643 |
- |
- |
PREDICTED: 11-beta-hydroxysteroid dehydrogenase-like 5 [Jatropha curcas] |