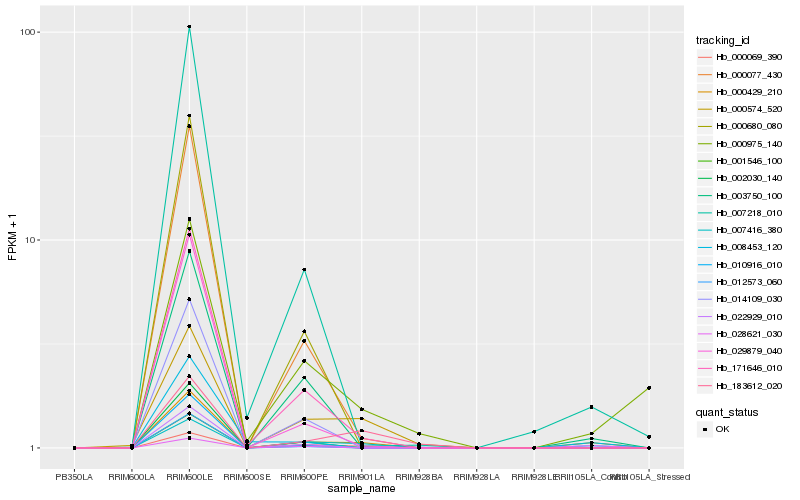
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000429_210 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_0089590 [Ricinus communis] |
| 2 |
Hb_007416_380 |
0.2084553994 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 3 [Jatropha curcas] |
| 3 |
Hb_171646_010 |
0.2093542883 |
- |
- |
hypothetical protein B456_012G144300, partial [Gossypium raimondii] |
| 4 |
Hb_000069_390 |
0.2106886988 |
- |
- |
PREDICTED: probable F-box protein At3g61730 [Jatropha curcas] |
| 5 |
Hb_000574_520 |
0.214862364 |
- |
- |
PREDICTED: uncharacterized protein LOC105634842 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003750_100 |
0.2181781703 |
- |
- |
HMGI/Y family protein [Populus trichocarpa] |
| 7 |
Hb_010916_010 |
0.2188045796 |
- |
- |
terpene synthase [Populus trichocarpa] |
| 8 |
Hb_007218_010 |
0.2257665249 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X1 [Populus euphratica] |
| 9 |
Hb_000077_430 |
0.226295363 |
- |
- |
hypothetical protein POPTR_0019s13390g [Populus trichocarpa] |
| 10 |
Hb_008453_120 |
0.2302065264 |
transcription factor |
TF Family: bZIP |
hypothetical protein POPTR_0005s26480g [Populus trichocarpa] |
| 11 |
Hb_183612_020 |
0.2359507777 |
- |
- |
PREDICTED: small heat shock protein, chloroplastic-like [Nelumbo nucifera] |
| 12 |
Hb_028621_030 |
0.2383519872 |
- |
- |
PREDICTED: serine/threonine-protein kinase AGC1-7 [Jatropha curcas] |
| 13 |
Hb_000975_140 |
0.2401635204 |
- |
- |
lactoylglutathione lyase family protein [Populus trichocarpa] |
| 14 |
Hb_029879_040 |
0.240862105 |
- |
- |
PREDICTED: cytochrome P450 77A4-like [Jatropha curcas] |
| 15 |
Hb_012573_060 |
0.2414792407 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 52C-like [Vitis vinifera] |
| 16 |
Hb_000680_080 |
0.2416390672 |
- |
- |
PREDICTED: protein HOTHEAD [Jatropha curcas] |
| 17 |
Hb_022929_010 |
0.2416633309 |
- |
- |
zinc/iron transporter, putative [Ricinus communis] |
| 18 |
Hb_002030_140 |
0.2417402286 |
- |
- |
PREDICTED: protein MIZU-KUSSEI 1-like [Jatropha curcas] |
| 19 |
Hb_014109_030 |
0.2421358129 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPEECHLESS [Jatropha curcas] |
| 20 |
Hb_001546_100 |
0.2421509682 |
transcription factor |
TF Family: AP2 |
hypothetical protein JCGZ_24704 [Jatropha curcas] |