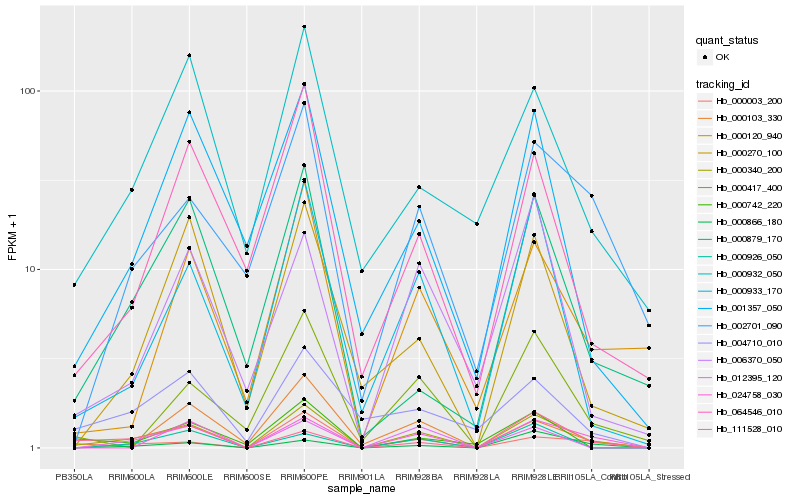
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000340_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105650039 [Jatropha curcas] |
| 2 |
Hb_000933_170 |
0.1990818609 |
- |
- |
PREDICTED: uncharacterized protein At5g01610 [Jatropha curcas] |
| 3 |
Hb_012395_120 |
0.2222925237 |
- |
- |
- |
| 4 |
Hb_024758_030 |
0.2288797676 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 5 |
Hb_001357_050 |
0.2317510813 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase 12-like [Jatropha curcas] |
| 6 |
Hb_000003_200 |
0.2350785971 |
- |
- |
hypothetical protein TRIUR3_35397 [Triticum urartu] |
| 7 |
Hb_000417_400 |
0.235339636 |
- |
- |
PREDICTED: ubiquitin domain-containing protein 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_064546_010 |
0.2367914676 |
- |
- |
PREDICTED: peroxidase 3 [Jatropha curcas] |
| 9 |
Hb_000270_100 |
0.2383131474 |
- |
- |
ATPase 4, plasma membrane-type -like protein [Gossypium arboreum] |
| 10 |
Hb_000742_220 |
0.2392473928 |
- |
- |
PREDICTED: uncharacterized protein LOC105633730 [Jatropha curcas] |
| 11 |
Hb_000103_330 |
0.2398683482 |
- |
- |
PREDICTED: meiotic recombination protein DMC1 homolog isoform X1 [Glycine max] |
| 12 |
Hb_006370_050 |
0.2402177544 |
- |
- |
Pectin lyase-like superfamily protein isoform 1 [Theobroma cacao] |
| 13 |
Hb_000879_170 |
0.2414049144 |
transcription factor |
TF Family: LIM |
PREDICTED: LIM domain-containing protein WLIM1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002701_090 |
0.2418897129 |
- |
- |
hypothetical protein POPTR_0017s13430g [Populus trichocarpa] |
| 15 |
Hb_000926_050 |
0.2428838921 |
- |
- |
PREDICTED: vinorine synthase-like [Populus euphratica] |
| 16 |
Hb_000932_050 |
0.2433768921 |
- |
- |
annexin [Manihot esculenta] |
| 17 |
Hb_111528_010 |
0.2435653905 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 2-like [Jatropha curcas] |
| 18 |
Hb_004710_010 |
0.2450702431 |
- |
- |
hypothetical protein EUGRSUZ_D010351, partial [Eucalyptus grandis] |
| 19 |
Hb_000120_940 |
0.2451783335 |
- |
- |
PREDICTED: phosphoserine aminotransferase 1, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_000866_180 |
0.2468064038 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |