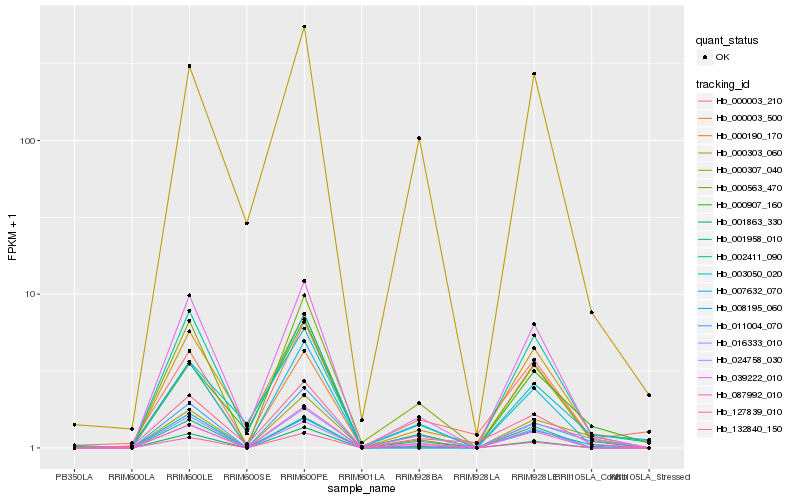
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000307_040 |
0.0 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 2 |
Hb_024758_030 |
0.1671900573 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 3 |
Hb_000907_160 |
0.1711704702 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 4 |
Hb_000563_470 |
0.1712900579 |
- |
- |
PREDICTED: uncharacterized protein LOC105643461 [Jatropha curcas] |
| 5 |
Hb_007632_070 |
0.17467816 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_039222_010 |
0.1776008403 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_132840_150 |
0.1872227803 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 8 |
Hb_000190_170 |
0.191533564 |
- |
- |
PREDICTED: chlorophyllase-1-like [Jatropha curcas] |
| 9 |
Hb_016333_010 |
0.1916043763 |
- |
- |
PREDICTED: probable terpene synthase 13 [Jatropha curcas] |
| 10 |
Hb_127839_010 |
0.1925205191 |
- |
- |
hypothetical protein PRUPE_ppa000956mg [Prunus persica] |
| 11 |
Hb_011004_070 |
0.1943339912 |
- |
- |
hypothetical protein RCOM_1494490 [Ricinus communis] |
| 12 |
Hb_087992_010 |
0.1964522131 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 13 |
Hb_000303_060 |
0.1999927646 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_001958_010 |
0.2023752087 |
- |
- |
hypothetical protein CICLE_v100078532mg, partial [Citrus clementina] |
| 15 |
Hb_001863_330 |
0.2030836193 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 16 |
Hb_003050_020 |
0.2033912952 |
- |
- |
glycosyl hydrolase family 17 family protein [Populus trichocarpa] |
| 17 |
Hb_002411_090 |
0.2044815397 |
- |
- |
PREDICTED: putative RING-H2 finger protein ATL69 [Jatropha curcas] |
| 18 |
Hb_000003_500 |
0.2058613878 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12-like [Jatropha curcas] |
| 19 |
Hb_000003_210 |
0.2074575281 |
- |
- |
PREDICTED: GEM-like protein 4 [Jatropha curcas] |
| 20 |
Hb_008195_060 |
0.2075171153 |
transcription factor |
TF Family: NAC |
NAC transcription factor 018 [Jatropha curcas] |