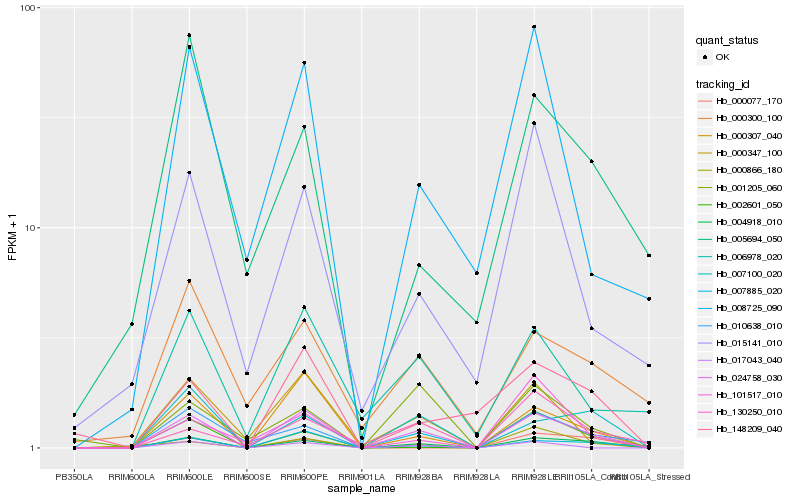
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004918_010 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_024758_030 |
0.1266782416 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 3 |
Hb_007100_020 |
0.1629319446 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 4 |
Hb_002601_050 |
0.2361691545 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 5 |
Hb_000307_040 |
0.2480629882 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 6 |
Hb_000077_170 |
0.2492810857 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Jatropha curcas] |
| 7 |
Hb_000866_180 |
0.2694295514 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 8 |
Hb_130250_010 |
0.2748642164 |
- |
- |
Putative disease resistance RGA4 [Gossypium arboreum] |
| 9 |
Hb_000300_100 |
0.2749780977 |
- |
- |
NADH-cytochrome b5 reductase 1 [Morus notabilis] |
| 10 |
Hb_010638_010 |
0.2757302655 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 11 |
Hb_007885_020 |
0.2760731213 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 12 |
Hb_148209_040 |
0.2797892608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001205_060 |
0.2800889207 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 14 |
Hb_005694_050 |
0.2830762992 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 15 |
Hb_006978_020 |
0.2868129478 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_000347_100 |
0.291870917 |
transcription factor |
TF Family: C3H |
hypothetical protein RCOM_0684740 [Ricinus communis] |
| 17 |
Hb_101517_010 |
0.2950508461 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 18 |
Hb_015141_010 |
0.2959786316 |
- |
- |
gamma-tocopherol methyltransferase [Hevea brasiliensis] |
| 19 |
Hb_008725_090 |
0.2963656898 |
- |
- |
PREDICTED: uncharacterized protein LOC105642581 [Jatropha curcas] |
| 20 |
Hb_017043_040 |
0.2984911816 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |