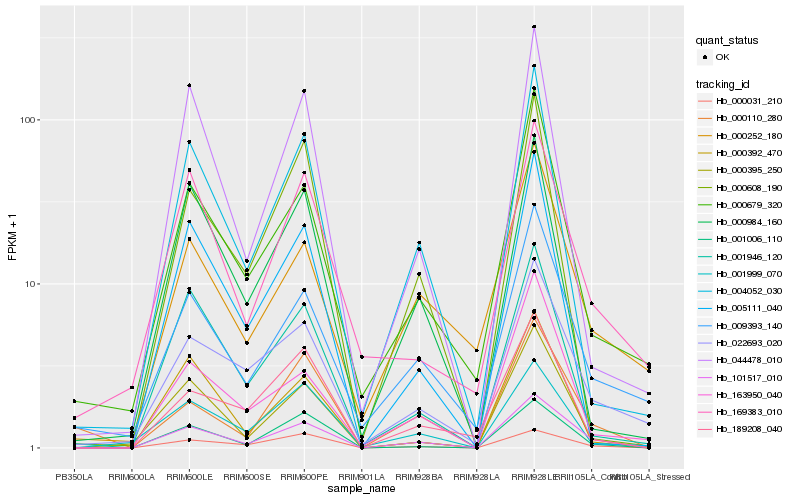
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101517_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 2 |
Hb_000395_250 |
0.1480802189 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 3 |
Hb_000031_210 |
0.1489513844 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_001006_110 |
0.1491721673 |
- |
- |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 5 |
Hb_022693_020 |
0.1564059642 |
- |
- |
pectin acetylesterase, putative [Ricinus communis] |
| 6 |
Hb_009393_140 |
0.1593461177 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004052_030 |
0.1642215041 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 8 |
Hb_000679_320 |
0.1677703541 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105136146 [Populus euphratica] |
| 9 |
Hb_044478_010 |
0.1736602733 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 10 |
Hb_163950_040 |
0.1766905393 |
- |
- |
PREDICTED: putative 4-hydroxy-tetrahydrodipicolinate reductase 3, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001946_120 |
0.1769862155 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 12 |
Hb_000392_470 |
0.1777154492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001999_070 |
0.177771226 |
- |
- |
PREDICTED: ammonium transporter 1 member 3 [Jatropha curcas] |
| 14 |
Hb_000110_280 |
0.1786061379 |
- |
- |
PREDICTED: uncharacterized protein LOC105642012, partial [Jatropha curcas] |
| 15 |
Hb_000252_180 |
0.1793165058 |
- |
- |
Glutamate--glyoxylate aminotransferase 2 [Morus notabilis] |
| 16 |
Hb_169383_010 |
0.1804197913 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005111_040 |
0.182443159 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 18 |
Hb_000984_160 |
0.1826888933 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_000608_190 |
0.1827994982 |
rubber biosynthesis |
Gene Name: Isopentenyl-diphosphate delta isomerase |
PREDICTED: isopentenyl-diphosphate Delta-isomerase I [Jatropha curcas] |
| 20 |
Hb_189208_040 |
0.1829008049 |
- |
- |
hypothetical protein POPTR_0002s18920g [Populus trichocarpa] |