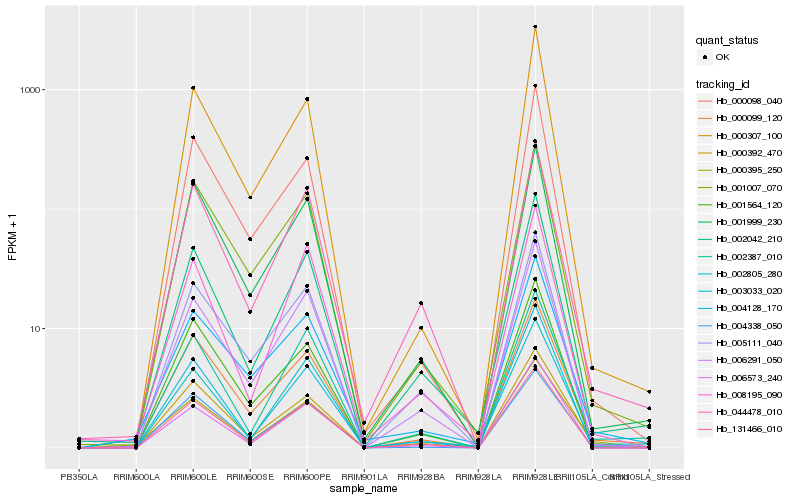
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000395_250 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 2 |
Hb_000392_470 |
0.083480965 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004128_170 |
0.0984227843 |
- |
- |
hypothetical protein POPTR_0016s08350g [Populus trichocarpa] |
| 4 |
Hb_002042_210 |
0.1026758591 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 5 |
Hb_006291_050 |
0.1035271117 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_000099_120 |
0.1052180895 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 7 |
Hb_044478_010 |
0.1060884913 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 8 |
Hb_131466_010 |
0.1123108685 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 9 |
Hb_002387_010 |
0.115232402 |
- |
- |
PREDICTED: protein STAY-GREEN LIKE, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002805_280 |
0.1170653166 |
- |
- |
PREDICTED: sugar transport protein 14-like [Jatropha curcas] |
| 11 |
Hb_005111_040 |
0.1183479736 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 12 |
Hb_001007_070 |
0.1193175975 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 13 |
Hb_004338_050 |
0.1198656673 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 14 |
Hb_008195_090 |
0.1206179495 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_001999_230 |
0.1208376547 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 16 |
Hb_001564_120 |
0.1214158159 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 17 |
Hb_000307_100 |
0.1223185792 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_006573_240 |
0.1233328597 |
- |
- |
PREDICTED: anthocyanidin 5,3-O-glucosyltransferase-like [Jatropha curcas] |
| 19 |
Hb_000098_040 |
0.1234307928 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_003033_020 |
0.1246493365 |
- |
- |
PREDICTED: glucomannan 4-beta-mannosyltransferase 9-like [Jatropha curcas] |