| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
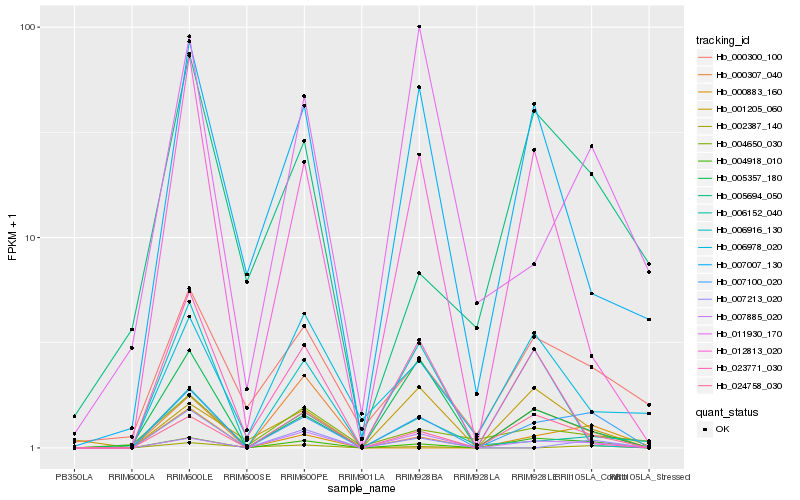
Hb_007100_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0473s00220g [Populus trichocarpa] |
| 2 |
Hb_004918_010 |
0.1629319446 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 3 |
Hb_024758_030 |
0.229010636 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 4 |
Hb_007885_020 |
0.2594930174 |
- |
- |
hypothetical protein JCGZ_04862 [Jatropha curcas] |
| 5 |
Hb_005357_180 |
0.2705859527 |
- |
- |
hypothetical protein POPTR_0011s00490g [Populus trichocarpa] |
| 6 |
Hb_000300_100 |
0.2770587247 |
- |
- |
NADH-cytochrome b5 reductase 1 [Morus notabilis] |
| 7 |
Hb_006152_040 |
0.2779872269 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105645062 isoform X2 [Jatropha curcas] |
| 8 |
Hb_012813_020 |
0.2826103459 |
- |
- |
PREDICTED: glucan endo-1,3-beta-glucosidase, basic isoform-like isoform X2 [Jatropha curcas] |
| 9 |
Hb_007007_130 |
0.2935271867 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 10 |
Hb_011930_170 |
0.301995079 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 11 |
Hb_004650_030 |
0.3036846849 |
- |
- |
PREDICTED: uncharacterized protein LOC105638888 isoform X1 [Jatropha curcas] |
| 12 |
Hb_023771_030 |
0.3067558198 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 13 |
Hb_006916_130 |
0.3101386259 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 14 |
Hb_007213_020 |
0.3103451769 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein At3g19184 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001205_060 |
0.3142406479 |
- |
- |
PREDICTED: protein N-lysine methyltransferase METTL21A [Jatropha curcas] |
| 16 |
Hb_000307_040 |
0.3155967968 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 17 |
Hb_000883_160 |
0.3168087623 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_005694_050 |
0.3180087962 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 19 |
Hb_002387_140 |
0.3240975238 |
- |
- |
PREDICTED: serine/threonine-protein kinase At5g01020-like [Jatropha curcas] |
| 20 |
Hb_006978_020 |
0.3254560028 |
- |
- |
PREDICTED: psbP domain-containing protein 5, chloroplastic isoform X2 [Jatropha curcas] |