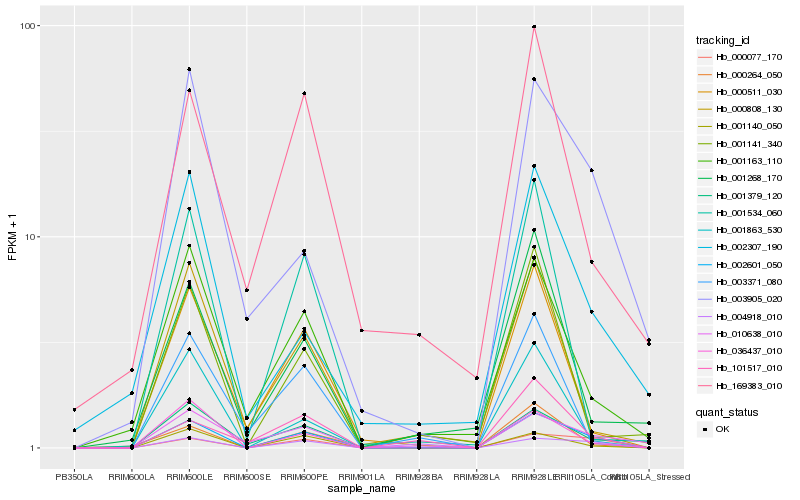
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002601_050 |
0.0 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 2 |
Hb_000077_170 |
0.1572794633 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Jatropha curcas] |
| 3 |
Hb_001140_050 |
0.1789432422 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1 [Jatropha curcas] |
| 4 |
Hb_036437_010 |
0.1889812835 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 5 |
Hb_001163_110 |
0.2083985823 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 6 |
Hb_000511_030 |
0.2148979274 |
- |
- |
PREDICTED: uncharacterized protein LOC105636463 [Jatropha curcas] |
| 7 |
Hb_010638_010 |
0.2157925013 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 8 |
Hb_000264_050 |
0.2167045194 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 9 |
Hb_001863_530 |
0.2233004907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000808_130 |
0.2244588643 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 11 |
Hb_003905_020 |
0.2266252687 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 12 |
Hb_001379_120 |
0.2266799144 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 13 |
Hb_101517_010 |
0.227908724 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 14 |
Hb_001141_340 |
0.2327191811 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 15 |
Hb_001534_060 |
0.2353684047 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C3 [Jatropha curcas] |
| 16 |
Hb_169383_010 |
0.2357175655 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003371_080 |
0.2360891659 |
- |
- |
hypothetical protein POPTR_0001s23660g [Populus trichocarpa] |
| 18 |
Hb_004918_010 |
0.2361691545 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_001268_170 |
0.2417176503 |
- |
- |
glycerophosphodiester phosphodiesterase, putative [Ricinus communis] |
| 20 |
Hb_002307_190 |
0.2419493116 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |