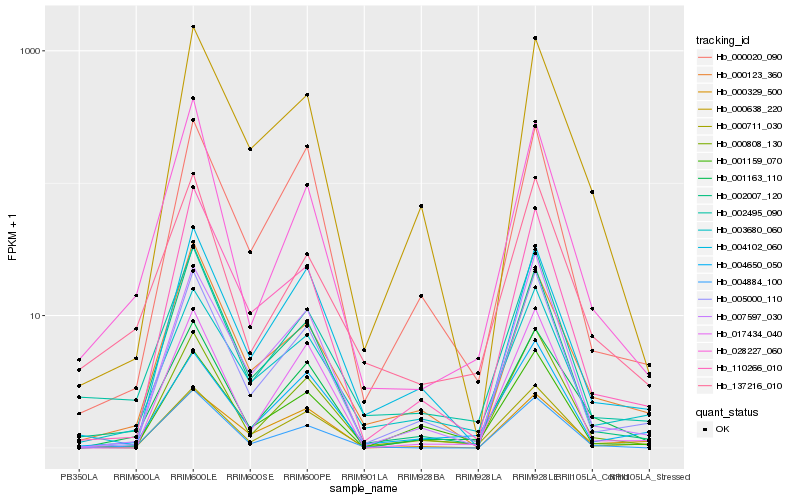
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001163_110 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 2 |
Hb_000808_130 |
0.1253196557 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |
| 3 |
Hb_005000_110 |
0.1330414673 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000711_030 |
0.1384785708 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 5 |
Hb_017434_040 |
0.1392920027 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670-like [Jatropha curcas] |
| 6 |
Hb_000638_220 |
0.1393231438 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000020_090 |
0.1426335934 |
- |
- |
hypothetical protein JCGZ_09084 [Jatropha curcas] |
| 8 |
Hb_028227_060 |
0.1429290396 |
- |
- |
PREDICTED: non-specific lipid transfer protein GPI-anchored 1-like [Populus euphratica] |
| 9 |
Hb_000329_500 |
0.1432746363 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 10 |
Hb_007597_030 |
0.14676548 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_003680_060 |
0.1468323854 |
- |
- |
hypothetical protein VITISV_022322 [Vitis vinifera] |
| 12 |
Hb_001159_070 |
0.14771954 |
- |
- |
DNA photolyase, putative [Ricinus communis] |
| 13 |
Hb_004884_100 |
0.1488176681 |
- |
- |
PREDICTED: lectin-domain containing receptor kinase VI.4-like [Jatropha curcas] |
| 14 |
Hb_137216_010 |
0.1497831237 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 15 |
Hb_004102_060 |
0.151848455 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 16 |
Hb_004650_050 |
0.153034223 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_000123_360 |
0.153124664 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_002007_120 |
0.1531791759 |
- |
- |
PREDICTED: psbP domain-containing protein 7, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002495_090 |
0.1541037639 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH48 isoform X2 [Jatropha curcas] |
| 20 |
Hb_110266_010 |
0.1549334993 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |