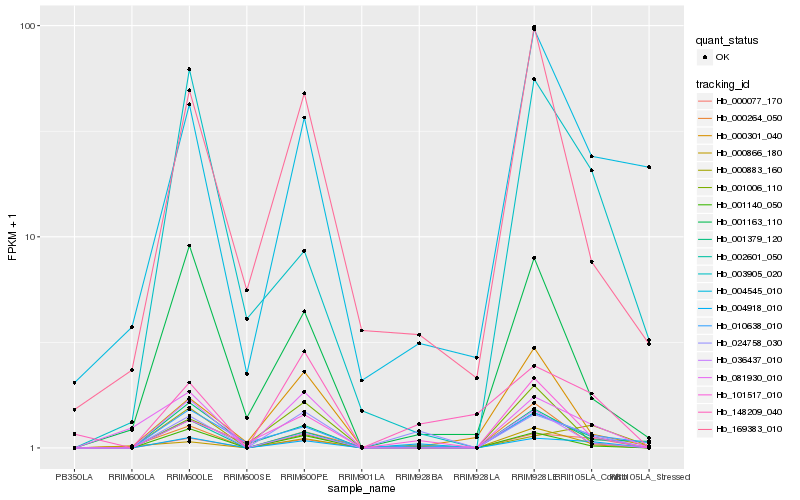
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_170 |
0.0 |
- |
- |
PREDICTED: GPI-anchored protein LORELEI-like [Jatropha curcas] |
| 2 |
Hb_002601_050 |
0.1572794633 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 3 |
Hb_004918_010 |
0.2492810857 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 4 |
Hb_081930_010 |
0.2692724178 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 5 |
Hb_001140_050 |
0.2723685821 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase EMS1 [Jatropha curcas] |
| 6 |
Hb_010638_010 |
0.274558239 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 7 |
Hb_003905_020 |
0.2900529275 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 8 |
Hb_000883_160 |
0.2926529933 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000866_180 |
0.2962647001 |
- |
- |
PREDICTED: feruloyl CoA ortho-hydroxylase 2-like [Populus euphratica] |
| 10 |
Hb_101517_010 |
0.2968871651 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 11 |
Hb_001379_120 |
0.3006383836 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 12 |
Hb_024758_030 |
0.3022316801 |
- |
- |
hypothetical protein JCGZ_16050 [Jatropha curcas] |
| 13 |
Hb_004545_010 |
0.3023457847 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 14 |
Hb_000264_050 |
0.307253166 |
- |
- |
hypothetical protein POPTR_0011s04750g [Populus trichocarpa] |
| 15 |
Hb_036437_010 |
0.3109690585 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 16 |
Hb_001006_110 |
0.3125607258 |
- |
- |
PREDICTED: uncharacterized protein LOC105634212 [Jatropha curcas] |
| 17 |
Hb_000301_040 |
0.3136606263 |
- |
- |
PREDICTED: uncharacterized protein LOC105639235 [Jatropha curcas] |
| 18 |
Hb_001163_110 |
0.3163916301 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 19 |
Hb_148209_040 |
0.3183305681 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_169383_010 |
0.3189070099 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |