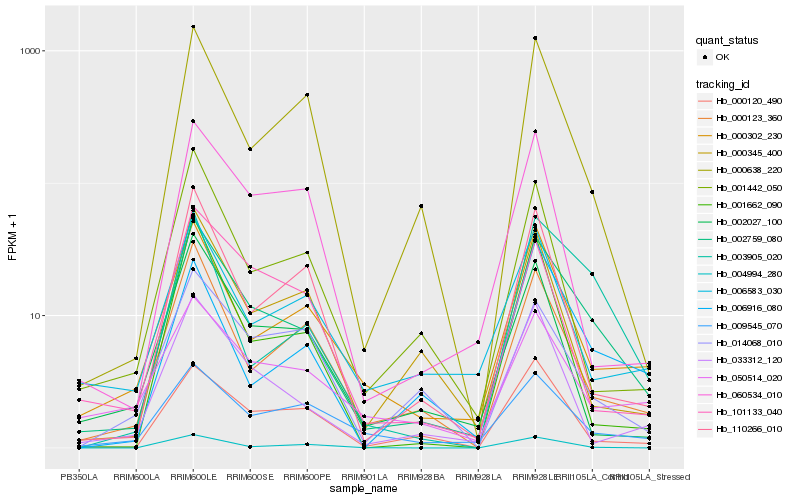
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002759_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 2 |
Hb_000302_230 |
0.166471369 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000123_360 |
0.1684411117 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_003905_020 |
0.1702711738 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 5 |
Hb_033312_120 |
0.1824585934 |
- |
- |
PREDICTED: psbQ-like protein 3, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000638_220 |
0.1833934643 |
- |
- |
PREDICTED: photosystem I reaction center subunit N, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000120_490 |
0.1878380067 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 8 |
Hb_009545_070 |
0.1879230428 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g14820 [Jatropha curcas] |
| 9 |
Hb_110266_010 |
0.1889477366 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g21222-like [Jatropha curcas] |
| 10 |
Hb_001442_050 |
0.1894825989 |
- |
- |
PREDICTED: NAD(P)H-quinone oxidoreductase subunit U, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002027_100 |
0.1906806017 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000345_400 |
0.1907475533 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_050514_020 |
0.1923516757 |
- |
- |
PREDICTED: abscisic acid receptor PYL4 [Jatropha curcas] |
| 14 |
Hb_101133_040 |
0.1935416453 |
- |
- |
PREDICTED: nudix hydrolase 18, mitochondrial-like [Jatropha curcas] |
| 15 |
Hb_006583_030 |
0.1936030164 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 16 |
Hb_060534_010 |
0.197530908 |
- |
- |
diphosphoinositol polyphosphate phosphohydrolase, putative [Ricinus communis] |
| 17 |
Hb_001662_090 |
0.1975351072 |
- |
- |
PREDICTED: thiosulfate sulfurtransferase 16, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_004994_280 |
0.1976478109 |
- |
- |
hypothetical protein JCGZ_14421 [Jatropha curcas] |
| 19 |
Hb_014068_010 |
0.1994818658 |
- |
- |
PREDICTED: phototropin-1 [Jatropha curcas] |
| 20 |
Hb_006916_080 |
0.2009101209 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |