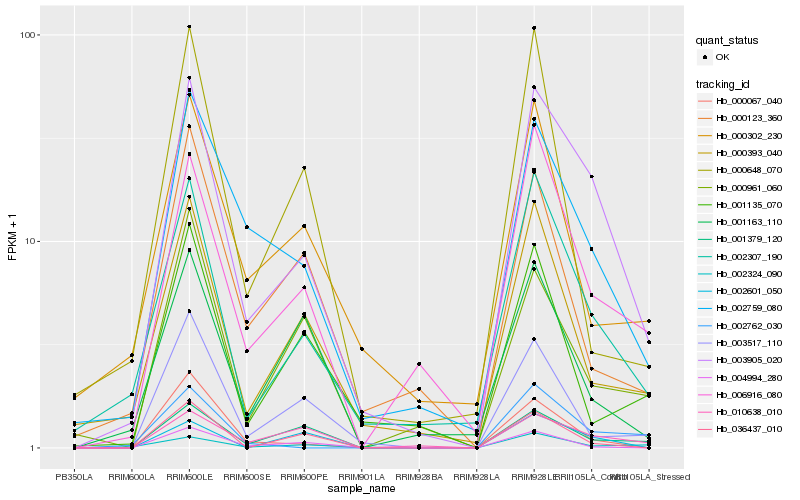
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003905_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 2 |
Hb_002307_190 |
0.1493977098 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 3 |
Hb_002759_080 |
0.1702711738 |
- |
- |
PREDICTED: uncharacterized protein LOC105649846 [Jatropha curcas] |
| 4 |
Hb_006916_080 |
0.1763798735 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 5 |
Hb_000393_040 |
0.2018650724 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 6 |
Hb_010638_010 |
0.2120608812 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 7 |
Hb_001379_120 |
0.213360658 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 8 |
Hb_000067_040 |
0.2158574152 |
- |
- |
PREDICTED: TMV resistance protein N-like [Jatropha curcas] |
| 9 |
Hb_002762_030 |
0.2182978743 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 10 |
Hb_003517_110 |
0.2202501451 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 11 |
Hb_000302_230 |
0.2257365922 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002601_050 |
0.2266252687 |
- |
- |
Boron transporter, putative [Ricinus communis] |
| 13 |
Hb_036437_010 |
0.233283426 |
transcription factor |
TF Family: bZIP |
PREDICTED: basic leucine zipper 61-like [Jatropha curcas] |
| 14 |
Hb_004994_280 |
0.2334739947 |
- |
- |
hypothetical protein JCGZ_14421 [Jatropha curcas] |
| 15 |
Hb_002324_090 |
0.2345133393 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 16 |
Hb_000123_360 |
0.2350370526 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_000961_060 |
0.2392051404 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 18 |
Hb_001163_110 |
0.2397108388 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 19 |
Hb_000648_070 |
0.2429032921 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 20 |
Hb_001135_070 |
0.2438571768 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |