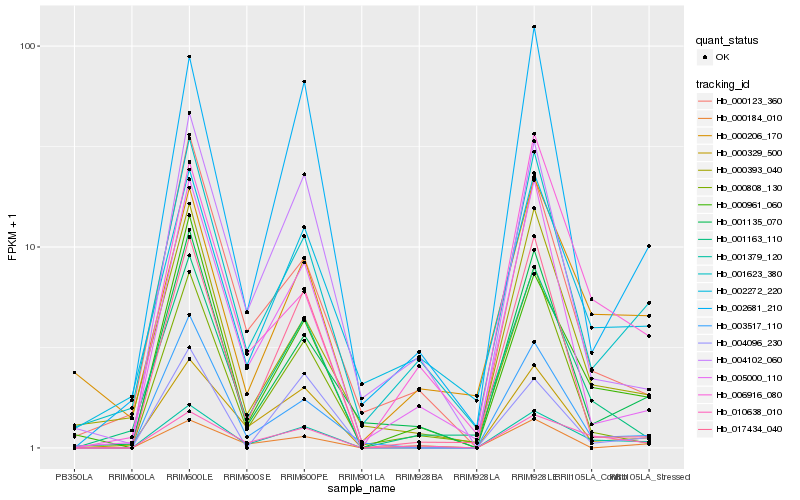
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001379_120 |
0.0 |
- |
- |
hypothetical protein POPTR_0001s29730g [Populus trichocarpa] |
| 2 |
Hb_010638_010 |
0.1184621975 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 3 |
Hb_000329_500 |
0.1497836667 |
- |
- |
PREDICTED: uncharacterized protein LOC105643180 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000961_060 |
0.1571022659 |
- |
- |
uvb-resistance protein uvr8, putative [Ricinus communis] |
| 5 |
Hb_003517_110 |
0.1608199235 |
- |
- |
PREDICTED: uncharacterized protein LOC105633058 [Jatropha curcas] |
| 6 |
Hb_001163_110 |
0.1674644516 |
- |
- |
PREDICTED: serine/threonine-protein kinase D6PKL2 [Jatropha curcas] |
| 7 |
Hb_001623_380 |
0.1724946686 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 8 |
Hb_000184_010 |
0.1847691807 |
- |
- |
mutt domain protein, putative [Ricinus communis] |
| 9 |
Hb_001135_070 |
0.1860309207 |
- |
- |
glutathione-s-transferase omega, putative [Ricinus communis] |
| 10 |
Hb_000393_040 |
0.1879244438 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 11 |
Hb_002272_220 |
0.1886625754 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002681_210 |
0.1910300615 |
- |
- |
PREDICTED: uncharacterized protein LOC105633686 [Jatropha curcas] |
| 13 |
Hb_004102_060 |
0.1926809029 |
- |
- |
JHL25H03.11 [Jatropha curcas] |
| 14 |
Hb_000206_170 |
0.1934773449 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 15 |
Hb_005000_110 |
0.1938263277 |
- |
- |
PREDICTED: uncharacterized protein LOC105637872 isoform X1 [Jatropha curcas] |
| 16 |
Hb_017434_040 |
0.1962228711 |
- |
- |
PREDICTED: uncharacterized oxidoreductase At4g09670-like [Jatropha curcas] |
| 17 |
Hb_000123_360 |
0.1974150972 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 1, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_004096_230 |
0.2000526732 |
- |
- |
PREDICTED: uncharacterized protein LOC105638704 isoform X1 [Jatropha curcas] |
| 19 |
Hb_006916_080 |
0.2021699799 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 20 |
Hb_000808_130 |
0.2030353961 |
- |
- |
3-5 exonuclease, putative [Ricinus communis] |