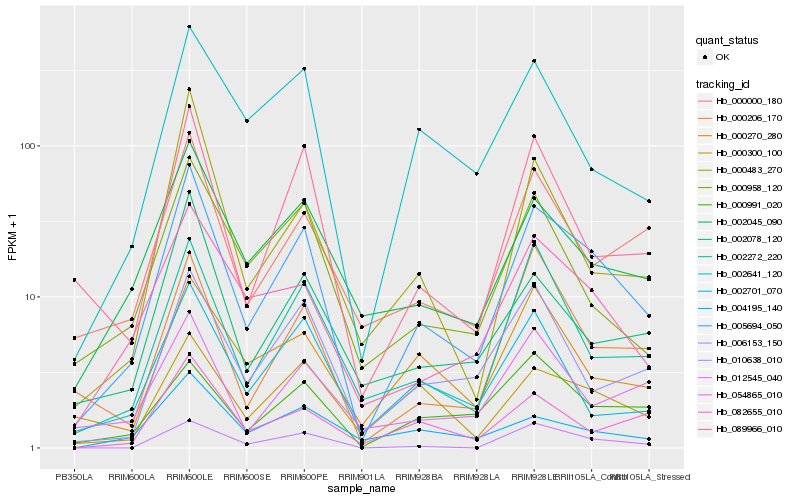
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005694_050 |
0.0 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 2 |
Hb_082655_010 |
0.1613586497 |
- |
- |
rhodanese-like domain-containing family protein [Populus trichocarpa] |
| 3 |
Hb_002045_090 |
0.1632758587 |
- |
- |
PREDICTED: tryptophan synthase alpha chain-like [Jatropha curcas] |
| 4 |
Hb_010638_010 |
0.1638778106 |
- |
- |
PREDICTED: RNA polymerase sigma factor sigC [Jatropha curcas] |
| 5 |
Hb_002272_220 |
0.1655014821 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_089966_010 |
0.1658252793 |
- |
- |
- |
| 7 |
Hb_000000_180 |
0.1718513774 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 8 |
Hb_002078_120 |
0.1792404945 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At1g67480 [Jatropha curcas] |
| 9 |
Hb_000958_120 |
0.1793039479 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 10 |
Hb_000300_100 |
0.180130929 |
- |
- |
NADH-cytochrome b5 reductase 1 [Morus notabilis] |
| 11 |
Hb_006153_150 |
0.1820428522 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 12 |
Hb_002641_120 |
0.1843582599 |
- |
- |
PREDICTED: calmodulin [Jatropha curcas] |
| 13 |
Hb_000206_170 |
0.188553033 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 14 |
Hb_002701_070 |
0.1893282185 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 15 |
Hb_054865_010 |
0.1895676146 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 16 |
Hb_000483_270 |
0.1910585602 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |
| 17 |
Hb_000270_280 |
0.1932476594 |
- |
- |
Rhicadhesin receptor precursor, putative [Ricinus communis] |
| 18 |
Hb_012545_040 |
0.1936296525 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 19 |
Hb_004195_140 |
0.1937704366 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 20 |
Hb_000991_020 |
0.1964230628 |
- |
- |
amino acid transporter, putative [Ricinus communis] |