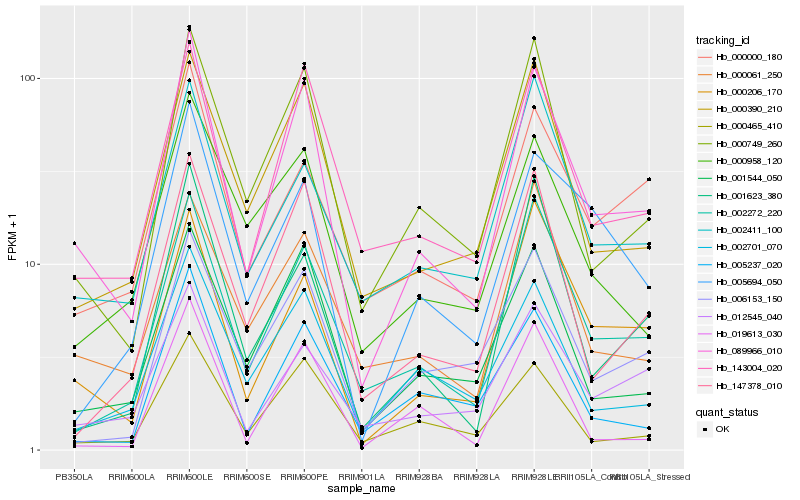
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_089966_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_143004_020 |
0.1127124121 |
- |
- |
PREDICTED: phosphopantothenate--cysteine ligase 2-like [Jatropha curcas] |
| 3 |
Hb_000749_260 |
0.1210433097 |
- |
- |
PREDICTED: protochlorophyllide reductase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001544_050 |
0.1320989426 |
- |
- |
PREDICTED: U-box domain-containing protein 11-like [Jatropha curcas] |
| 5 |
Hb_000390_210 |
0.1349743402 |
- |
- |
cinnamoyl-CoA reductase, putative [Ricinus communis] |
| 6 |
Hb_002272_220 |
0.1355047209 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000465_410 |
0.1365944094 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_147378_010 |
0.1421718045 |
- |
- |
- |
| 9 |
Hb_000206_170 |
0.1458857092 |
- |
- |
hypothetical protein JCGZ_25514 [Jatropha curcas] |
| 10 |
Hb_002701_070 |
0.1478539492 |
- |
- |
PREDICTED: uncharacterized protein LOC105646760 [Jatropha curcas] |
| 11 |
Hb_005237_020 |
0.1530633875 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 12 |
Hb_006153_150 |
0.1596532117 |
- |
- |
PREDICTED: uncharacterized protein LOC105645999 [Jatropha curcas] |
| 13 |
Hb_012545_040 |
0.1605932962 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_000061_250 |
0.1609419925 |
- |
- |
hypothetical protein JCGZ_11665 [Jatropha curcas] |
| 15 |
Hb_000958_120 |
0.1625647352 |
- |
- |
PREDICTED: uncharacterized protein LOC105646385 [Jatropha curcas] |
| 16 |
Hb_001623_380 |
0.1641900595 |
- |
- |
PREDICTED: 7-hydroxymethyl chlorophyll a reductase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_005694_050 |
0.1658252793 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 18 |
Hb_000000_180 |
0.1664266738 |
- |
- |
PREDICTED: 30S ribosomal protein S9, chloroplastic-like [Populus euphratica] |
| 19 |
Hb_019613_030 |
0.1668343321 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 20 |
Hb_002411_100 |
0.1675453866 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |