| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
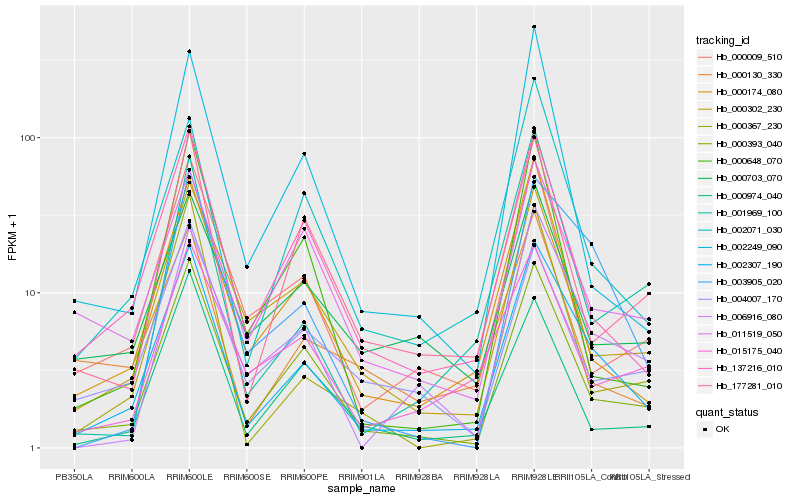
Hb_002307_190 |
0.0 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 2 |
Hb_000393_040 |
0.1185635752 |
- |
- |
PREDICTED: uncharacterized protein LOC105128208 [Populus euphratica] |
| 3 |
Hb_000174_080 |
0.1421895997 |
- |
- |
4-nitrophenylphosphatase, putative [Ricinus communis] |
| 4 |
Hb_137216_010 |
0.1430967699 |
- |
- |
PREDICTED: unknown protein DS12 from 2D-PAGE of leaf, chloroplastic-like isoform X2 [Citrus sinensis] |
| 5 |
Hb_002071_030 |
0.1457937539 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 6 |
Hb_003905_020 |
0.1493977098 |
- |
- |
PREDICTED: uncharacterized protein LOC105131878 isoform X1 [Populus euphratica] |
| 7 |
Hb_006916_080 |
0.1526734086 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |
| 8 |
Hb_000302_230 |
0.1593377471 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000009_510 |
0.1622158916 |
- |
- |
PREDICTED: uncharacterized protein LOC105126496 [Populus euphratica] |
| 10 |
Hb_000367_230 |
0.1724344906 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 11 |
Hb_002249_090 |
0.1729658699 |
- |
- |
PREDICTED: magnesium protoporphyrin IX methyltransferase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_177281_010 |
0.1769664476 |
- |
- |
PREDICTED: 50S ribosomal protein 5, chloroplastic-like [Citrus sinensis] |
| 13 |
Hb_015175_040 |
0.1794419046 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000130_330 |
0.1798988029 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP3-like [Jatropha curcas] |
| 15 |
Hb_000648_070 |
0.1813697529 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 16 |
Hb_000974_040 |
0.1818548603 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Jatropha curcas] |
| 17 |
Hb_001969_100 |
0.1826741873 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 18 |
Hb_011519_050 |
0.1856145822 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |
| 19 |
Hb_004007_170 |
0.1859727953 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 20 |
Hb_000703_070 |
0.1862532137 |
- |
- |
PREDICTED: uncharacterized protein LOC105635704 isoform X1 [Jatropha curcas] |