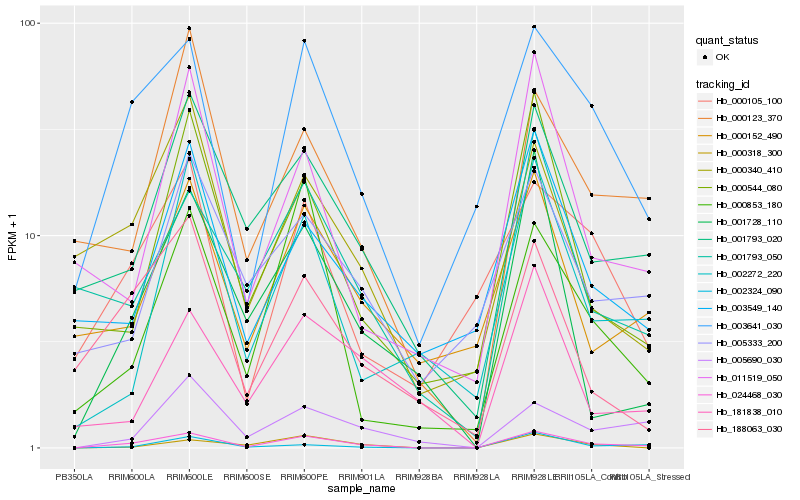
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_024468_030 |
0.0 |
desease resistance |
Gene Name: DUF3388 |
putative disease resistance gene NBS-LRR family protein [Populus trichocarpa] |
| 2 |
Hb_003641_030 |
0.185317866 |
- |
- |
PREDICTED: uncharacterized protein LOC105638015 [Jatropha curcas] |
| 3 |
Hb_001793_020 |
0.2064645228 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 4 |
Hb_005690_030 |
0.2169367497 |
- |
- |
hypothetical protein VITISV_020378 [Vitis vinifera] |
| 5 |
Hb_002324_090 |
0.2234977692 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 6 |
Hb_001728_110 |
0.2263416807 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF039-like [Jatropha curcas] |
| 7 |
Hb_000318_300 |
0.2295790942 |
transcription factor |
TF Family: B3 |
PREDICTED: uncharacterized protein LOC105634339 [Jatropha curcas] |
| 8 |
Hb_001793_050 |
0.2310740054 |
- |
- |
PREDICTED: uncharacterized protein LOC105632663 [Jatropha curcas] |
| 9 |
Hb_002272_220 |
0.2330637326 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000123_370 |
0.2334674416 |
- |
- |
PREDICTED: psbP-like protein 1, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000544_080 |
0.2336630977 |
- |
- |
PREDICTED: uncharacterized protein LOC105631062 isoform X3 [Jatropha curcas] |
| 12 |
Hb_000853_180 |
0.2347151604 |
- |
- |
kinesin heavy chain, putative [Ricinus communis] |
| 13 |
Hb_000152_490 |
0.2353410659 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 14 |
Hb_188063_030 |
0.2373700846 |
- |
- |
PREDICTED: protein LONGIFOLIA 1 [Jatropha curcas] |
| 15 |
Hb_003549_140 |
0.2419942587 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000340_410 |
0.242552209 |
- |
- |
PREDICTED: receptor-like protein 12 [Jatropha curcas] |
| 17 |
Hb_000105_100 |
0.2428547267 |
- |
- |
PREDICTED: pyruvate kinase isozyme A, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_181838_010 |
0.2433996511 |
- |
- |
PREDICTED: uncharacterized protein LOC105631275 [Jatropha curcas] |
| 19 |
Hb_005333_200 |
0.2449747231 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_011519_050 |
0.2453156437 |
- |
- |
hypothetical protein POPTR_0015s04810g [Populus trichocarpa] |