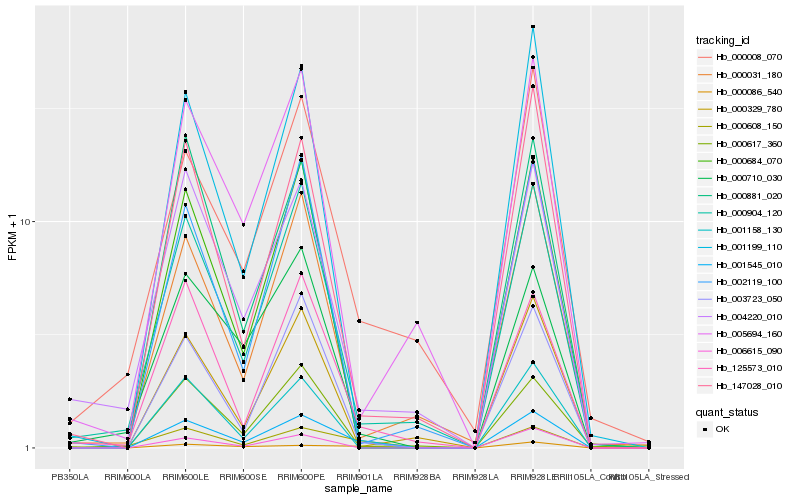
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001545_010 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase TNNI3K-like [Cucumis sativus] |
| 2 |
Hb_000904_120 |
0.1448625348 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003723_050 |
0.1481589085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_125573_010 |
0.1483521988 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_000684_070 |
0.16085086 |
- |
- |
PREDICTED: uncharacterized protein LOC105642198 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000608_150 |
0.1610728085 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_147028_010 |
0.1636967201 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein HD1 [Prunus mume] |
| 8 |
Hb_001158_130 |
0.1637305841 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 9 |
Hb_000710_030 |
0.1656110402 |
- |
- |
PREDICTED: uncharacterized protein LOC105629456 [Jatropha curcas] |
| 10 |
Hb_000008_070 |
0.1672929574 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_004220_010 |
0.1673356622 |
- |
- |
PREDICTED: uncharacterized protein LOC105642366 [Jatropha curcas] |
| 12 |
Hb_000617_360 |
0.1682132833 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001199_110 |
0.1687979409 |
- |
- |
PREDICTED: uncharacterized protein ycf36 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000031_180 |
0.1718402189 |
- |
- |
sarcosine oxidase, putative [Ricinus communis] |
| 15 |
Hb_002119_100 |
0.1722702399 |
- |
- |
cytochrome B561, putative [Ricinus communis] |
| 16 |
Hb_000329_780 |
0.1724469354 |
- |
- |
hypothetical protein POPTR_0403s00200g [Populus trichocarpa] |
| 17 |
Hb_005694_160 |
0.1725825704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_006615_090 |
0.1730036672 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 19 |
Hb_000881_020 |
0.1760666197 |
- |
- |
hypothetical protein MIMGU_mgv1a022110mg, partial [Erythranthe guttata] |
| 20 |
Hb_000086_540 |
0.176203139 |
- |
- |
- |